Gene
KWMTBOMO01377
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.371
Sequence
CDS
ATGTATGGCAGCGAAACATGGGCAGCAACCAAAAGGAACGTACATACCATTCAAGTTGCTGAAATGAAAATGCTGAGGTGGATGTGCGGCGTGACTAGGCTCGATAAAATCCGCAACGAGTACGTGCGTGGAAGTCTAGGTGTACGGGACATCGCCGACAAGATGCAGGAGAGTAGGCTGAGATGGTATGGTCACGTGAAAAGGAAGCCACCTGACTACGTCGGAAATTTGGCGCTACTGCTCGACCTCCCCGGTCGGAGACCTAGAGGCAGACCCAAGACAAGGTGGAGGGACGTAGTGCTGAAGGACAAGAGAGAATGCAATGTTGCTGACGAGAATGTCGAAGACAGAGCGAAGTGGAGGAGTAAAGTCAGGAAAGCTGACCCCACCACCATGTGGGATAAATAG
Protein
MYGSETWAATKRNVHTIQVAEMKMLRWMCGVTRLDKIRNEYVRGSLGVRDIADKMQESRLRWYGHVKRKPPDYVGNLALLLDLPGRRPRGRPKTRWRDVVLKDKRECNVADENVEDRAKWRSKVRKADPTTMWDK
Summary
Uniprot
D7F169
C6Y4D5
A0A183FXH7
A0A3P7Z2B4
K1KXQ8
A0A183G637
+ More
A0A3P8A2N8 I7K8L3 A0A2I0B7E5 A0A016TSY7 A0A016TKX6 A0A0N4WFX2 A0A2I0A7U6 A0A016TK27 A0A2H9ZV89 A0A2I0A8C5 A0A016TQJ7 G0XZC9 A0A016W910 A0A2G9U5Q2 A0A016TLV5 A0A016T5F5 A0A016VVS4 A0A016WHD5 A0A2I0A7B2 A0A2I0BDY2 A0A2I0BFB2 A0A2H9ZWL9 A0A3B3HQB6 A0A3P9MAU2 A0A016VT97 A0A183GPW8 A0A3P8ETQ0 A0A3P9K1J1 A0A2I0A4R0 A0A3B3HF10 A0A2G9U478 A0A183GFD9 A0A3P8CQ14 A0A2I0A8G9 A0A3P9JDN6 A0A016V9U6 A0A3B3H8Q9 A0A3B3HJS0 A0A3P9JAC7 A0A016UI69 A0A3P9M3G7 H2MG20 A0A183FSE4 A0A3P7Z8X4 A0A3P9I3Z4 A0A3B3HFD6 A0A183FT77 A0A3P8AAR6 A0A016VCI1 A0A183GGD8 A0A3P8D1Q5 A0A183G8L9 A0A3P8BMV8 A0A3P9IJG8 W6NPY2 A0A183F4B8 A0A3P7UF10 A0A183F2R5 A0A3P7U320 A0A3B3HF03 A0A3B3IFI4 A0A016RVL3 A0A183G962 A0A3P8BZL1 A0A3B3I2L1 A0A183F3R1 A0A3P7TDA1 A0A3B3IMV3 A0A3B3IC01 A0A3P9HQ45 A0A3P9K727 A0A016SDV3 W6NHZ4 A0A3B3HGK1 A0A016VW07 A0A016V285 A0A016RTP4 A0A183FB99 A0A3P7VL99 W6NF39 A0A3P9HCZ5 A0A183GA74 A0A3P8F0W9 A0A2T7DJ34 A0A3P9HH58 A0A016THU0 A0A016SG79 A0A3B3H692 A0A3P9IV37 A0A183G741 A0A3P8A7C9 A0A016SBW0 A0A3P9K097 A0A3P9MDZ4
A0A3P8A2N8 I7K8L3 A0A2I0B7E5 A0A016TSY7 A0A016TKX6 A0A0N4WFX2 A0A2I0A7U6 A0A016TK27 A0A2H9ZV89 A0A2I0A8C5 A0A016TQJ7 G0XZC9 A0A016W910 A0A2G9U5Q2 A0A016TLV5 A0A016T5F5 A0A016VVS4 A0A016WHD5 A0A2I0A7B2 A0A2I0BDY2 A0A2I0BFB2 A0A2H9ZWL9 A0A3B3HQB6 A0A3P9MAU2 A0A016VT97 A0A183GPW8 A0A3P8ETQ0 A0A3P9K1J1 A0A2I0A4R0 A0A3B3HF10 A0A2G9U478 A0A183GFD9 A0A3P8CQ14 A0A2I0A8G9 A0A3P9JDN6 A0A016V9U6 A0A3B3H8Q9 A0A3B3HJS0 A0A3P9JAC7 A0A016UI69 A0A3P9M3G7 H2MG20 A0A183FSE4 A0A3P7Z8X4 A0A3P9I3Z4 A0A3B3HFD6 A0A183FT77 A0A3P8AAR6 A0A016VCI1 A0A183GGD8 A0A3P8D1Q5 A0A183G8L9 A0A3P8BMV8 A0A3P9IJG8 W6NPY2 A0A183F4B8 A0A3P7UF10 A0A183F2R5 A0A3P7U320 A0A3B3HF03 A0A3B3IFI4 A0A016RVL3 A0A183G962 A0A3P8BZL1 A0A3B3I2L1 A0A183F3R1 A0A3P7TDA1 A0A3B3IMV3 A0A3B3IC01 A0A3P9HQ45 A0A3P9K727 A0A016SDV3 W6NHZ4 A0A3B3HGK1 A0A016VW07 A0A016V285 A0A016RTP4 A0A183FB99 A0A3P7VL99 W6NF39 A0A3P9HCZ5 A0A183GA74 A0A3P8F0W9 A0A2T7DJ34 A0A3P9HH58 A0A016THU0 A0A016SG79 A0A3B3H692 A0A3P9IV37 A0A183G741 A0A3P8A7C9 A0A016SBW0 A0A3P9K097 A0A3P9MDZ4
EMBL
FJ265554
ADI61822.1
CU462842
CBA11992.1
UZAH01027842
VDO95476.1
+ More
AMGM01000184 EKB47241.1 UZAH01029822 VDP08057.1 HE805490 CCH50976.1 KZ451907 PKA63681.1 JARK01001415 EYC05905.1 JARK01001430 EYC03360.1 UZAF01001415 UZAF01017114 VDO08263.1 VDO38063.1 KZ452013 PKA51585.1 EYC03359.1 KZ453539 PKA47214.1 KZ452012 PKA51786.1 JARK01001422 EYC04723.1 HQ445899 AEJ72569.1 JARK01000539 EYC36061.1 KZ348915 PIO65553.1 JARK01001428 EYC03715.1 JARK01001472 EYB97834.1 JARK01001340 EYC31495.1 JARK01000281 EYC39011.1 PKA51430.1 KZ451888 PKA66013.1 KZ451885 PKA66495.1 KZ453102 PKA47709.1 JARK01001341 EYC30277.1 UZAH01036764 VDP46995.1 KZ452023 PKA50536.1 KZ350014 PIO64310.1 UZAH01032701 VDP23368.1 PKA51832.1 JARK01001350 EYC24439.1 JARK01001375 EYC14890.1 UZAH01026905 VDO86609.1 UZAH01027034 VDO87933.1 EYC24438.1 UZAH01033088 VDP26174.1 UZAH01030549 VDP10928.1 CAVP010057772 CDL94282.1 UZAH01000951 VDO19393.1 UZAH01000306 VDO18824.1 JARK01001702 EYB82127.1 UZAH01030698 VDP11665.1 UZAH01000694 VDO19186.1 JARK01001582 EYB88469.1 CAVP010062000 CDL96883.1 EYC31496.1 JARK01001356 EYC21107.1 JARK01001722 JARK01001346 EYB81349.1 EYC26203.1 UZAH01009099 VDO35114.1 CAVP010059598 CDL95931.1 UZAH01030983 VDP13266.1 CM009753 PUZ55583.1 JARK01001437 EYC02250.1 JARK01001564 EYB89718.1 UZAH01030101 VDP09262.1 JARK01001592 EYB87852.1
AMGM01000184 EKB47241.1 UZAH01029822 VDP08057.1 HE805490 CCH50976.1 KZ451907 PKA63681.1 JARK01001415 EYC05905.1 JARK01001430 EYC03360.1 UZAF01001415 UZAF01017114 VDO08263.1 VDO38063.1 KZ452013 PKA51585.1 EYC03359.1 KZ453539 PKA47214.1 KZ452012 PKA51786.1 JARK01001422 EYC04723.1 HQ445899 AEJ72569.1 JARK01000539 EYC36061.1 KZ348915 PIO65553.1 JARK01001428 EYC03715.1 JARK01001472 EYB97834.1 JARK01001340 EYC31495.1 JARK01000281 EYC39011.1 PKA51430.1 KZ451888 PKA66013.1 KZ451885 PKA66495.1 KZ453102 PKA47709.1 JARK01001341 EYC30277.1 UZAH01036764 VDP46995.1 KZ452023 PKA50536.1 KZ350014 PIO64310.1 UZAH01032701 VDP23368.1 PKA51832.1 JARK01001350 EYC24439.1 JARK01001375 EYC14890.1 UZAH01026905 VDO86609.1 UZAH01027034 VDO87933.1 EYC24438.1 UZAH01033088 VDP26174.1 UZAH01030549 VDP10928.1 CAVP010057772 CDL94282.1 UZAH01000951 VDO19393.1 UZAH01000306 VDO18824.1 JARK01001702 EYB82127.1 UZAH01030698 VDP11665.1 UZAH01000694 VDO19186.1 JARK01001582 EYB88469.1 CAVP010062000 CDL96883.1 EYC31496.1 JARK01001356 EYC21107.1 JARK01001722 JARK01001346 EYB81349.1 EYC26203.1 UZAH01009099 VDO35114.1 CAVP010059598 CDL95931.1 UZAH01030983 VDP13266.1 CM009753 PUZ55583.1 JARK01001437 EYC02250.1 JARK01001564 EYB89718.1 UZAH01030101 VDP09262.1 JARK01001592 EYB87852.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F169
C6Y4D5
A0A183FXH7
A0A3P7Z2B4
K1KXQ8
A0A183G637
+ More
A0A3P8A2N8 I7K8L3 A0A2I0B7E5 A0A016TSY7 A0A016TKX6 A0A0N4WFX2 A0A2I0A7U6 A0A016TK27 A0A2H9ZV89 A0A2I0A8C5 A0A016TQJ7 G0XZC9 A0A016W910 A0A2G9U5Q2 A0A016TLV5 A0A016T5F5 A0A016VVS4 A0A016WHD5 A0A2I0A7B2 A0A2I0BDY2 A0A2I0BFB2 A0A2H9ZWL9 A0A3B3HQB6 A0A3P9MAU2 A0A016VT97 A0A183GPW8 A0A3P8ETQ0 A0A3P9K1J1 A0A2I0A4R0 A0A3B3HF10 A0A2G9U478 A0A183GFD9 A0A3P8CQ14 A0A2I0A8G9 A0A3P9JDN6 A0A016V9U6 A0A3B3H8Q9 A0A3B3HJS0 A0A3P9JAC7 A0A016UI69 A0A3P9M3G7 H2MG20 A0A183FSE4 A0A3P7Z8X4 A0A3P9I3Z4 A0A3B3HFD6 A0A183FT77 A0A3P8AAR6 A0A016VCI1 A0A183GGD8 A0A3P8D1Q5 A0A183G8L9 A0A3P8BMV8 A0A3P9IJG8 W6NPY2 A0A183F4B8 A0A3P7UF10 A0A183F2R5 A0A3P7U320 A0A3B3HF03 A0A3B3IFI4 A0A016RVL3 A0A183G962 A0A3P8BZL1 A0A3B3I2L1 A0A183F3R1 A0A3P7TDA1 A0A3B3IMV3 A0A3B3IC01 A0A3P9HQ45 A0A3P9K727 A0A016SDV3 W6NHZ4 A0A3B3HGK1 A0A016VW07 A0A016V285 A0A016RTP4 A0A183FB99 A0A3P7VL99 W6NF39 A0A3P9HCZ5 A0A183GA74 A0A3P8F0W9 A0A2T7DJ34 A0A3P9HH58 A0A016THU0 A0A016SG79 A0A3B3H692 A0A3P9IV37 A0A183G741 A0A3P8A7C9 A0A016SBW0 A0A3P9K097 A0A3P9MDZ4
A0A3P8A2N8 I7K8L3 A0A2I0B7E5 A0A016TSY7 A0A016TKX6 A0A0N4WFX2 A0A2I0A7U6 A0A016TK27 A0A2H9ZV89 A0A2I0A8C5 A0A016TQJ7 G0XZC9 A0A016W910 A0A2G9U5Q2 A0A016TLV5 A0A016T5F5 A0A016VVS4 A0A016WHD5 A0A2I0A7B2 A0A2I0BDY2 A0A2I0BFB2 A0A2H9ZWL9 A0A3B3HQB6 A0A3P9MAU2 A0A016VT97 A0A183GPW8 A0A3P8ETQ0 A0A3P9K1J1 A0A2I0A4R0 A0A3B3HF10 A0A2G9U478 A0A183GFD9 A0A3P8CQ14 A0A2I0A8G9 A0A3P9JDN6 A0A016V9U6 A0A3B3H8Q9 A0A3B3HJS0 A0A3P9JAC7 A0A016UI69 A0A3P9M3G7 H2MG20 A0A183FSE4 A0A3P7Z8X4 A0A3P9I3Z4 A0A3B3HFD6 A0A183FT77 A0A3P8AAR6 A0A016VCI1 A0A183GGD8 A0A3P8D1Q5 A0A183G8L9 A0A3P8BMV8 A0A3P9IJG8 W6NPY2 A0A183F4B8 A0A3P7UF10 A0A183F2R5 A0A3P7U320 A0A3B3HF03 A0A3B3IFI4 A0A016RVL3 A0A183G962 A0A3P8BZL1 A0A3B3I2L1 A0A183F3R1 A0A3P7TDA1 A0A3B3IMV3 A0A3B3IC01 A0A3P9HQ45 A0A3P9K727 A0A016SDV3 W6NHZ4 A0A3B3HGK1 A0A016VW07 A0A016V285 A0A016RTP4 A0A183FB99 A0A3P7VL99 W6NF39 A0A3P9HCZ5 A0A183GA74 A0A3P8F0W9 A0A2T7DJ34 A0A3P9HH58 A0A016THU0 A0A016SG79 A0A3B3H692 A0A3P9IV37 A0A183G741 A0A3P8A7C9 A0A016SBW0 A0A3P9K097 A0A3P9MDZ4
Ontologies
KEGG
PANTHER
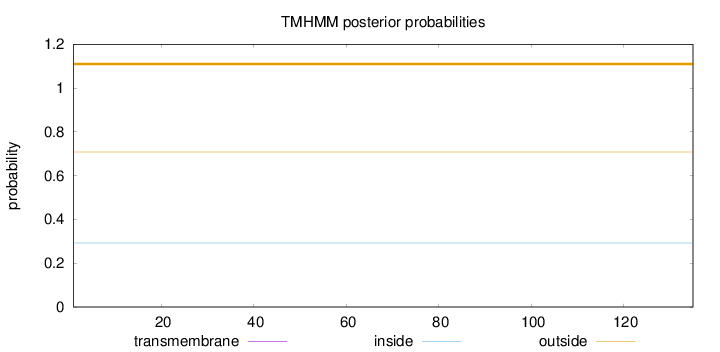
Topology
Length:
135
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00189
Exp number, first 60 AAs:
0.00154
Total prob of N-in:
0.29253
outside
1 - 135
Population Genetic Test Statistics
Pi
243.572614
Theta
192.895349
Tajima's D
0.815471
CLR
0
CSRT
0.604319784010799
Interpretation
Uncertain
