Gene
KWMTBOMO01376 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008986
Annotation
integrin_beta-nu_precursor_[Bombyx_mori]
Full name
Integrin beta
+ More
Integrin beta-nu
Integrin beta-nu
Location in the cell
Extracellular Reliability : 2.852
Sequence
CDS
ATGTACGATTATACGTTGTGTAAATTGTTAGTGTTTTCAGTTATTATAATATACACGTGCGGTCAAGTTCAAGAAAAACTGCTCAACAAGCTAGTGTGCATCGAGCATGAAGAGTGCGGTGCGTGTTTGTCGGCTGCTTCGCACTGTCGATGGTGTGCAGACCCTTACTTCAGTCACGCTGCTCCCAGGTGCAACGACGACGAAAGTTTGGTGTCTACGGGATGCAGTCAAGCTATGATACAACGACCAGAGAAACCTATATGGGTTGTGGCCGAGAATAGTTCCTTACAAGACATGCAACCGGACAGTGAAGAGTCCGTAGTACAGATCCAGCCTCAAAGGATCAAATTATCGTTGAAACCACGTGAAACAAAAAAAATTAAGTTCGTATACCGGCCAGCGAAGAATTACCCTCTAGATCTCTACTACCTGATGGACTTGACCTGGTCAATGAAGGACGACAAGGAAACACTGGTTTCCTTAAGGGATGACTTGCCTATTCTGTTAAAGAATTTAACTGACAATTTTAGGCTCGGTTTTGGTAGTTTTGCTGATAAACCAATCATGCCATTCATCAACGTGGATGATAGAAGAAAATCAAACCCATGTGCTGTTGAGGAAGAAGCTTGCGAGGCAACATACAGCTACAAACACCATTTATCTCTTACTAATGAGGTAAGCGATTTTATCAAGAAAGTAAACACGAGTTCAGTGACTGCAAATCTGGATAACGCCGAGGCCCAATTAGATGCCCTCATGCAGGTCTTGACTTGCGGCGAGAAGATCGGATGGTCGACGAAAAGCAGAAAGATCGTAATTCTCTCTTCAGATGGACTGTTGCATATTGCTGGCGACGGAAGGATAGGGGGAGCAGCATTAAAGAACGATGAAAATTGCCATCTGGATGACAACGGCTACTATAACAAGGCGGACACCTACGATTATCCATCTATTGCTCAAATCTTTAGATTACTTGATAAGTATAAGGTTAACGTTATATTTGCTGTAACCGAAAGTGTTAAGGATCATTACGATAGTCTACACGAACTTTTAAAAGATTTCACGTACGTAGCTAAATTGGAAAGTGACAGTTCCAACATTCTAAAGCTCGTCAAAACGGGATACGAAGACATCGTCAGCGTTGTCGATTTTGAGGACAATGCAAGCTCGGGCCCGGCTAAGGTTCGGTACTTCTCTGATTGTGGAGTGAAAGGAGGGTCGCTTGTCGAGACGACGCGATGTACCGGAGTCGAATACGGCATGACCCTCAATTACGAGGCGCACATCACTCTCGATGCTTGTTCGGAATTTAAAACTTCGAGTCAGACGATTCGCATTTCCGAAAGTCAACTTGGACAGGACTCGCTCACTCTCGATGTGGAATTACAATGTGGATGCAAATGCGAAGGCGATTCGCAGAAAGATCTGTCCCTGACGTGCCCAGTCAATTCTCACCTCGTGTGCGGTGTTTGTCAGTGTAATAAAGGCTGGTCCGGACCAGACTGTGACTGTTCAATAAGCGACGAGGAAGCATCTGCGGAACTAATGGCTCAATGTCGAGAGCCGAACTCGACGCGGACTCTCGCCTGTTCCGGCGCCGGAGACTGCATCTGCGGGAGGTGCGACTGCGACCGTGGCTACAACGGAAAATATTGCCAGTGCAAGTCGTGTGAAACTAGTCCAGAAAACGGTATCGAATGTGGCGGTGTGGGTCGCGGCGTGTGTGTTTGTGGACAATGTGCGTGTGTCGACGGCTGGGGCGGACCTTCCTGTGACTGCACCACCGTATTGGATTCGTGTATTGCACCGGGAGACGAAAAACTTTGTTCCGGACATGGAGACTGTGTTTGTGGTCAGTGTCAGTGTTCCGCCTCTGATCAGAGCAGCGGCTCCGTATACTCAGGCACGTTTTGCGAGACATGTGCAACCTGTGAGAATCCGCTATGCGCAAACGCGGAACCCTGCGTTTCTTGTCATCTCAACAGCAGTTGTACGGATATATGTACGATTGGCAGCGTCAACTACACAGTCCATGACAGAATGAGCGAAACTGGGTCCGTAAGCGCGGAAGATATCACCTGTATCCTTCGCATAGAGGAGAACTCACTTGAATGCGAATACAAATACACTTACAACGCGGTCAGACAGTCCATAGTCGCCATGGAGATTGCGCTCCGTTCTAAAGACTGCTTCCAGCCGACCAGCGCAAAGATCATGACCAGCGCTTTCGTCATCATAGCCTGCGTGATAGCGGCAGGGCTTCTAATTATCTTAGCCATAAAGAGCGCTCAGATCGTTTCAGACAGGCGAGCATACGCGAAATTCGTTAAAGAAGCACAGGAGAGTCGAAAGAATATGCAAGAACTGAACCCGTTGTATAAATCTCCTATAACAGAGTTTAAGCTGCCGGATTCGTTCCCTAGAGATAAGAATGATTAA
Protein
MYDYTLCKLLVFSVIIIYTCGQVQEKLLNKLVCIEHEECGACLSAASHCRWCADPYFSHAAPRCNDDESLVSTGCSQAMIQRPEKPIWVVAENSSLQDMQPDSEESVVQIQPQRIKLSLKPRETKKIKFVYRPAKNYPLDLYYLMDLTWSMKDDKETLVSLRDDLPILLKNLTDNFRLGFGSFADKPIMPFINVDDRRKSNPCAVEEEACEATYSYKHHLSLTNEVSDFIKKVNTSSVTANLDNAEAQLDALMQVLTCGEKIGWSTKSRKIVILSSDGLLHIAGDGRIGGAALKNDENCHLDDNGYYNKADTYDYPSIAQIFRLLDKYKVNVIFAVTESVKDHYDSLHELLKDFTYVAKLESDSSNILKLVKTGYEDIVSVVDFEDNASSGPAKVRYFSDCGVKGGSLVETTRCTGVEYGMTLNYEAHITLDACSEFKTSSQTIRISESQLGQDSLTLDVELQCGCKCEGDSQKDLSLTCPVNSHLVCGVCQCNKGWSGPDCDCSISDEEASAELMAQCREPNSTRTLACSGAGDCICGRCDCDRGYNGKYCQCKSCETSPENGIECGGVGRGVCVCGQCACVDGWGGPSCDCTTVLDSCIAPGDEKLCSGHGDCVCGQCQCSASDQSSGSVYSGTFCETCATCENPLCANAEPCVSCHLNSSCTDICTIGSVNYTVHDRMSETGSVSAEDITCILRIEENSLECEYKYTYNAVRQSIVAMEIALRSKDCFQPTSAKIMTSAFVIIACVIAAGLLIILAIKSAQIVSDRRAYAKFVKEAQESRKNMQELNPLYKSPITEFKLPDSFPRDKND
Summary
Description
Contributes to endodermal integrity and adhesion between the midgut epithelium and the surrounding visceral muscle. Essential for migration of the primordial midgut cells and for maintaining, but not establishing, cell polarity in the midgut epithelium. Can only partially compensate for the loss of beta-PS integrin during primordial midgut cell migration. The two beta subunits mediate midgut migration by distinct mechanisms: beta-PS requires rhea/Talin and beta-nu does not. Integrin alpha-PS3/beta-nu is required for effective phagocytosis of apoptotic cells during embryonic development and for the phagocytic elimination of S.aureus by mediating the binding of S.aureus peptidoglycan to larval hemocytes, which probably activates a signaling pathway involving Rac1 and Rac2. Not required for the production of antimicrobial peptides during S.aureus. Upon activation by LanA, integrin alpha-PS3/beta-nu activates Fak in presynapsis to suppress neuromuscular junction (NMJ) growth during larval development and during low crawling activity, but not during higher-crawling conditions. Mediates, together with LanA, glutamate receptor-modulated NMJ growth.
Subunit
Heterodimer of an alpha and a beta subunit. Interacts with scb/alpha-PS3.
Similarity
Belongs to the integrin beta chain family.
Keywords
Cell adhesion
Complete proteome
Glycoprotein
Integrin
Membrane
Phagocytosis
Receptor
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Integrin beta
Uniprot
A0A076JUD9
H9JHI8
A0A194RMR4
A0A194PW01
A0A2A4KA02
A0A212FDP2
+ More
A0A2H1WGU6 A0A1E1VXT8 A0A0L7L1Q4 A0A2W1B938 A0A1W7R6Y7 A0A1Q3FQ99 A0A1Q3FQF2 A0A336LEA7 A0A336KAI0 A0A1Q3FQA7 A0A182R6X4 A0A182VSW8 A0A3L8D5H4 Q16TB6 A0A1L8DLQ6 A0A1L8DLZ9 A0A1L8DLQ7 B0W9T9 A0A310SFX4 A0A182JW23 Q17K64 E2BNA6 A0A182Q580 A0A182P887 A0A0N0U352 A0A1A9ZPW0 A0A1A9V8Z9 A0A1A9Y5S3 Q7PME7 Q8T5U7 A0A182KJT3 A0A182I0S3 A0A182XM86 A0A1J1HHK0 A0A0L0C681 A0A088A188 W5J9X8 A0A182N8S2 A0A0L7R5I8 A0A2A3ELY3 A0A1W4UI89 A0A182GK62 B4MZU0 A0A1B0DC03 A0A1I8M0W6 B4KL00 A0A1B0DL74 B3MU64 A0A182FMW5 B4JEA5 B4IFJ1 T1P8K2 A0A3B0JIT6 Q29NR4 B4GJH0 A0A3B0K923 A0A0S0WGT1 Q27591 A0A0J9R574 A0A1B0B8X4 B4LUM4 B4P709 B3NKV9 A0A1I8P121 A0A0R1DSU0 A0A1W7RAE7 T1K5U0 V5IFB6 A0A0R3NXK3 A0A026VXQ7 A0A2I9LP51 V5GLP5 A0A1Y3EQP6 A0A1I8P153 A0A0M3QUB4 F0JAH6 A0A2M4BDW1 A0A0A0PEU5 D3K4F4 A0A2M4BDV9 A0A1S4FW80 A0A182Q0X5 A0A182RHJ6 T1GNV7 A0A1I7RZ20 B0WJW0 A0A336M8L1 A0A336MBS4 U5EVW7 A0A0K0ELJ0 T1JH92 A0A0N5D3K5 A0A1B0FKA3
A0A2H1WGU6 A0A1E1VXT8 A0A0L7L1Q4 A0A2W1B938 A0A1W7R6Y7 A0A1Q3FQ99 A0A1Q3FQF2 A0A336LEA7 A0A336KAI0 A0A1Q3FQA7 A0A182R6X4 A0A182VSW8 A0A3L8D5H4 Q16TB6 A0A1L8DLQ6 A0A1L8DLZ9 A0A1L8DLQ7 B0W9T9 A0A310SFX4 A0A182JW23 Q17K64 E2BNA6 A0A182Q580 A0A182P887 A0A0N0U352 A0A1A9ZPW0 A0A1A9V8Z9 A0A1A9Y5S3 Q7PME7 Q8T5U7 A0A182KJT3 A0A182I0S3 A0A182XM86 A0A1J1HHK0 A0A0L0C681 A0A088A188 W5J9X8 A0A182N8S2 A0A0L7R5I8 A0A2A3ELY3 A0A1W4UI89 A0A182GK62 B4MZU0 A0A1B0DC03 A0A1I8M0W6 B4KL00 A0A1B0DL74 B3MU64 A0A182FMW5 B4JEA5 B4IFJ1 T1P8K2 A0A3B0JIT6 Q29NR4 B4GJH0 A0A3B0K923 A0A0S0WGT1 Q27591 A0A0J9R574 A0A1B0B8X4 B4LUM4 B4P709 B3NKV9 A0A1I8P121 A0A0R1DSU0 A0A1W7RAE7 T1K5U0 V5IFB6 A0A0R3NXK3 A0A026VXQ7 A0A2I9LP51 V5GLP5 A0A1Y3EQP6 A0A1I8P153 A0A0M3QUB4 F0JAH6 A0A2M4BDW1 A0A0A0PEU5 D3K4F4 A0A2M4BDV9 A0A1S4FW80 A0A182Q0X5 A0A182RHJ6 T1GNV7 A0A1I7RZ20 B0WJW0 A0A336M8L1 A0A336MBS4 U5EVW7 A0A0K0ELJ0 T1JH92 A0A0N5D3K5 A0A1B0FKA3
Pubmed
25064490
19121390
26354079
22118469
26227816
28756777
+ More
30249741 17510324 20798317 12364791 14747013 17210077 20966253 26108605 20920257 23761445 26483478 17994087 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8076521 15469969 18925939 21592968 22547074 23054837 23426364 22936249 17550304 25765539 24508170 29248469 24982879 21909270
30249741 17510324 20798317 12364791 14747013 17210077 20966253 26108605 20920257 23761445 26483478 17994087 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8076521 15469969 18925939 21592968 22547074 23054837 23426364 22936249 17550304 25765539 24508170 29248469 24982879 21909270
EMBL
KJ511858
AII79417.1
BABH01012588
KQ459989
KPJ18714.1
KQ459591
+ More
KPI96939.1 NWSH01000002 PCG81051.1 AGBW02009016 OWR51852.1 ODYU01008576 SOQ52295.1 GDQN01011508 JAT79546.1 JTDY01003666 KOB69184.1 KZ150259 PZC71768.1 GEHC01000731 JAV46914.1 GFDL01005270 JAV29775.1 GFDL01005281 JAV29764.1 UFQS01003657 UFQT01003657 SSX15829.1 SSX35175.1 UFQS01000170 UFQT01000170 SSX00698.1 SSX21078.1 GFDL01005256 JAV29789.1 QOIP01000013 RLU15411.1 CH477653 EAT37755.1 GFDF01006686 JAV07398.1 GFDF01006687 JAV07397.1 GFDF01006688 JAV07396.1 DS231866 EDS40506.1 KQ764881 OAD54370.1 CH477228 EAT47028.1 GL449385 EFN82868.1 AXCN02001862 KQ435908 KOX68994.1 AAAB01008980 EAA13939.2 AF492464 AAM11657.1 APCN01005251 CVRI01000004 CRK87503.1 JRES01000842 KNC27795.1 ADMH02002068 ETN59660.1 KQ414652 KOC66084.1 KZ288215 PBC32717.1 JXUM01013004 KQ560366 KXJ82792.1 CH963920 EDW77875.2 AJVK01030471 AJVK01030472 CH933807 EDW12750.2 AJVK01036185 AJVK01036186 CH902624 EDV33393.2 CH916368 EDW03625.1 CH480833 EDW46413.1 KA644460 AFP59089.1 OUUW01000006 SPP82125.1 CH379058 EAL34580.3 CH479184 EDW36786.1 SPP82126.1 AE014134 ALI30216.1 L13305 BT031252 CM002910 KMY91263.1 JXJN01010098 CH940649 EDW64210.1 CM000158 EDW89978.2 CH954179 EDV54482.1 KRJ98850.1 GFAH01000303 JAV48086.1 CAEY01001590 GANP01008722 JAB75746.1 KRT03680.1 KK107796 EZA47614.1 GFWZ01000163 MBW20153.1 GANP01013203 JAB71265.1 LVZM01008211 OUC45859.1 CP012523 ALC40339.1 BT126018 ADY17717.1 GGFJ01002056 MBW51197.1 KC715739 AHH32890.1 GU370128 ADC35398.1 GGFJ01002095 MBW51236.1 AXCN02002227 CAQQ02145466 CAQQ02145467 DS231965 EDS29490.1 UFQT01000673 SSX26380.1 SSX26379.1 GANO01001694 JAB58177.1 JH432223 UYYF01004515 VDN04972.1 CCAG010009532
KPI96939.1 NWSH01000002 PCG81051.1 AGBW02009016 OWR51852.1 ODYU01008576 SOQ52295.1 GDQN01011508 JAT79546.1 JTDY01003666 KOB69184.1 KZ150259 PZC71768.1 GEHC01000731 JAV46914.1 GFDL01005270 JAV29775.1 GFDL01005281 JAV29764.1 UFQS01003657 UFQT01003657 SSX15829.1 SSX35175.1 UFQS01000170 UFQT01000170 SSX00698.1 SSX21078.1 GFDL01005256 JAV29789.1 QOIP01000013 RLU15411.1 CH477653 EAT37755.1 GFDF01006686 JAV07398.1 GFDF01006687 JAV07397.1 GFDF01006688 JAV07396.1 DS231866 EDS40506.1 KQ764881 OAD54370.1 CH477228 EAT47028.1 GL449385 EFN82868.1 AXCN02001862 KQ435908 KOX68994.1 AAAB01008980 EAA13939.2 AF492464 AAM11657.1 APCN01005251 CVRI01000004 CRK87503.1 JRES01000842 KNC27795.1 ADMH02002068 ETN59660.1 KQ414652 KOC66084.1 KZ288215 PBC32717.1 JXUM01013004 KQ560366 KXJ82792.1 CH963920 EDW77875.2 AJVK01030471 AJVK01030472 CH933807 EDW12750.2 AJVK01036185 AJVK01036186 CH902624 EDV33393.2 CH916368 EDW03625.1 CH480833 EDW46413.1 KA644460 AFP59089.1 OUUW01000006 SPP82125.1 CH379058 EAL34580.3 CH479184 EDW36786.1 SPP82126.1 AE014134 ALI30216.1 L13305 BT031252 CM002910 KMY91263.1 JXJN01010098 CH940649 EDW64210.1 CM000158 EDW89978.2 CH954179 EDV54482.1 KRJ98850.1 GFAH01000303 JAV48086.1 CAEY01001590 GANP01008722 JAB75746.1 KRT03680.1 KK107796 EZA47614.1 GFWZ01000163 MBW20153.1 GANP01013203 JAB71265.1 LVZM01008211 OUC45859.1 CP012523 ALC40339.1 BT126018 ADY17717.1 GGFJ01002056 MBW51197.1 KC715739 AHH32890.1 GU370128 ADC35398.1 GGFJ01002095 MBW51236.1 AXCN02002227 CAQQ02145466 CAQQ02145467 DS231965 EDS29490.1 UFQT01000673 SSX26380.1 SSX26379.1 GANO01001694 JAB58177.1 JH432223 UYYF01004515 VDN04972.1 CCAG010009532
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000075900 UP000075920 UP000279307 UP000008820 UP000002320 UP000075881 UP000008237 UP000075886 UP000075885 UP000053105 UP000092445 UP000078200 UP000092443 UP000007062 UP000075882 UP000075840 UP000076407 UP000183832 UP000037069 UP000005203 UP000000673 UP000075884 UP000053825 UP000242457 UP000192221 UP000069940 UP000249989 UP000007798 UP000092462 UP000095301 UP000009192 UP000007801 UP000069272 UP000001070 UP000001292 UP000268350 UP000001819 UP000008744 UP000000803 UP000092460 UP000008792 UP000002282 UP000008711 UP000095300 UP000015104 UP000053097 UP000243006 UP000092553 UP000015102 UP000095284 UP000035681 UP000046394 UP000276776 UP000092444
UP000075900 UP000075920 UP000279307 UP000008820 UP000002320 UP000075881 UP000008237 UP000075886 UP000075885 UP000053105 UP000092445 UP000078200 UP000092443 UP000007062 UP000075882 UP000075840 UP000076407 UP000183832 UP000037069 UP000005203 UP000000673 UP000075884 UP000053825 UP000242457 UP000192221 UP000069940 UP000249989 UP000007798 UP000092462 UP000095301 UP000009192 UP000007801 UP000069272 UP000001070 UP000001292 UP000268350 UP000001819 UP000008744 UP000000803 UP000092460 UP000008792 UP000002282 UP000008711 UP000095300 UP000015104 UP000053097 UP000243006 UP000092553 UP000015102 UP000095284 UP000035681 UP000046394 UP000276776 UP000092444
Pfam
Interpro
IPR036465
vWFA_dom_sf
+ More
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR002369 Integrin_bsu_VWA
IPR013111 EGF_extracell
IPR002035 VWF_A
IPR032695 Integrin_dom_sf
IPR033760 Integrin_beta_N
IPR040622 I-EGF_1
IPR000742 EGF-like_dom
IPR036349 Integrin_bsu_tail_dom_sf
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR016201 PSI
IPR012896 Integrin_bsu_tail
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR002369 Integrin_bsu_VWA
IPR013111 EGF_extracell
IPR002035 VWF_A
IPR032695 Integrin_dom_sf
IPR033760 Integrin_beta_N
IPR040622 I-EGF_1
IPR000742 EGF-like_dom
IPR036349 Integrin_bsu_tail_dom_sf
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR016201 PSI
IPR012896 Integrin_bsu_tail
Gene 3D
ProteinModelPortal
A0A076JUD9
H9JHI8
A0A194RMR4
A0A194PW01
A0A2A4KA02
A0A212FDP2
+ More
A0A2H1WGU6 A0A1E1VXT8 A0A0L7L1Q4 A0A2W1B938 A0A1W7R6Y7 A0A1Q3FQ99 A0A1Q3FQF2 A0A336LEA7 A0A336KAI0 A0A1Q3FQA7 A0A182R6X4 A0A182VSW8 A0A3L8D5H4 Q16TB6 A0A1L8DLQ6 A0A1L8DLZ9 A0A1L8DLQ7 B0W9T9 A0A310SFX4 A0A182JW23 Q17K64 E2BNA6 A0A182Q580 A0A182P887 A0A0N0U352 A0A1A9ZPW0 A0A1A9V8Z9 A0A1A9Y5S3 Q7PME7 Q8T5U7 A0A182KJT3 A0A182I0S3 A0A182XM86 A0A1J1HHK0 A0A0L0C681 A0A088A188 W5J9X8 A0A182N8S2 A0A0L7R5I8 A0A2A3ELY3 A0A1W4UI89 A0A182GK62 B4MZU0 A0A1B0DC03 A0A1I8M0W6 B4KL00 A0A1B0DL74 B3MU64 A0A182FMW5 B4JEA5 B4IFJ1 T1P8K2 A0A3B0JIT6 Q29NR4 B4GJH0 A0A3B0K923 A0A0S0WGT1 Q27591 A0A0J9R574 A0A1B0B8X4 B4LUM4 B4P709 B3NKV9 A0A1I8P121 A0A0R1DSU0 A0A1W7RAE7 T1K5U0 V5IFB6 A0A0R3NXK3 A0A026VXQ7 A0A2I9LP51 V5GLP5 A0A1Y3EQP6 A0A1I8P153 A0A0M3QUB4 F0JAH6 A0A2M4BDW1 A0A0A0PEU5 D3K4F4 A0A2M4BDV9 A0A1S4FW80 A0A182Q0X5 A0A182RHJ6 T1GNV7 A0A1I7RZ20 B0WJW0 A0A336M8L1 A0A336MBS4 U5EVW7 A0A0K0ELJ0 T1JH92 A0A0N5D3K5 A0A1B0FKA3
A0A2H1WGU6 A0A1E1VXT8 A0A0L7L1Q4 A0A2W1B938 A0A1W7R6Y7 A0A1Q3FQ99 A0A1Q3FQF2 A0A336LEA7 A0A336KAI0 A0A1Q3FQA7 A0A182R6X4 A0A182VSW8 A0A3L8D5H4 Q16TB6 A0A1L8DLQ6 A0A1L8DLZ9 A0A1L8DLQ7 B0W9T9 A0A310SFX4 A0A182JW23 Q17K64 E2BNA6 A0A182Q580 A0A182P887 A0A0N0U352 A0A1A9ZPW0 A0A1A9V8Z9 A0A1A9Y5S3 Q7PME7 Q8T5U7 A0A182KJT3 A0A182I0S3 A0A182XM86 A0A1J1HHK0 A0A0L0C681 A0A088A188 W5J9X8 A0A182N8S2 A0A0L7R5I8 A0A2A3ELY3 A0A1W4UI89 A0A182GK62 B4MZU0 A0A1B0DC03 A0A1I8M0W6 B4KL00 A0A1B0DL74 B3MU64 A0A182FMW5 B4JEA5 B4IFJ1 T1P8K2 A0A3B0JIT6 Q29NR4 B4GJH0 A0A3B0K923 A0A0S0WGT1 Q27591 A0A0J9R574 A0A1B0B8X4 B4LUM4 B4P709 B3NKV9 A0A1I8P121 A0A0R1DSU0 A0A1W7RAE7 T1K5U0 V5IFB6 A0A0R3NXK3 A0A026VXQ7 A0A2I9LP51 V5GLP5 A0A1Y3EQP6 A0A1I8P153 A0A0M3QUB4 F0JAH6 A0A2M4BDW1 A0A0A0PEU5 D3K4F4 A0A2M4BDV9 A0A1S4FW80 A0A182Q0X5 A0A182RHJ6 T1GNV7 A0A1I7RZ20 B0WJW0 A0A336M8L1 A0A336MBS4 U5EVW7 A0A0K0ELJ0 T1JH92 A0A0N5D3K5 A0A1B0FKA3
PDB
4UM8
E-value=3.96406e-85,
Score=805
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 21,
Likelihood: 0.867072
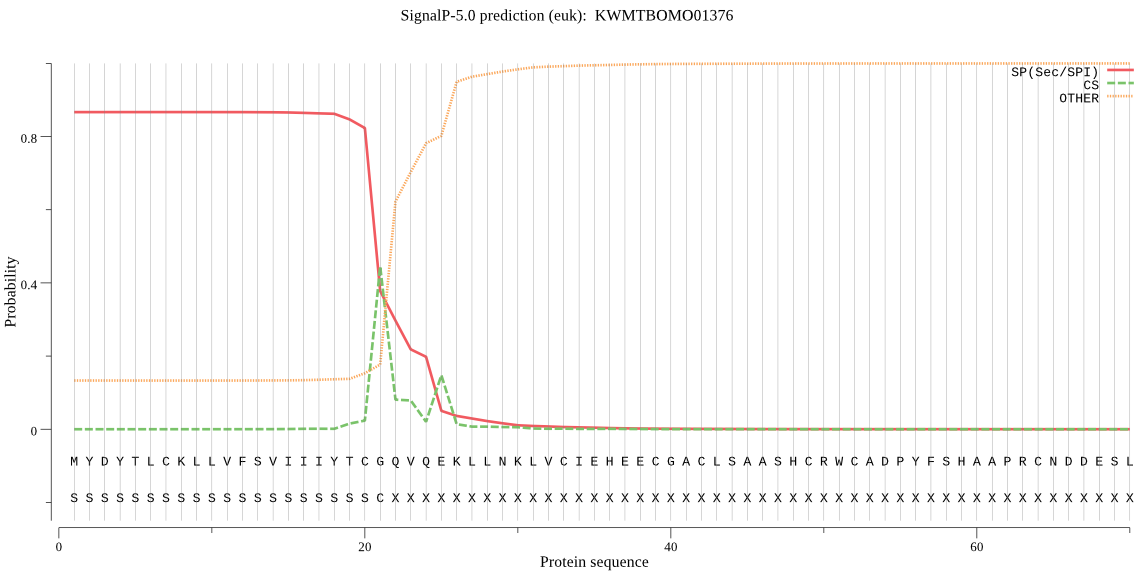
Length:
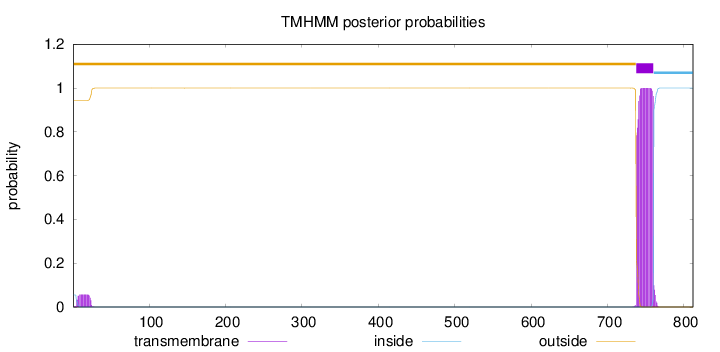
812
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.6481
Exp number, first 60 AAs:
1.04842
Total prob of N-in:
0.05703
outside
1 - 737
TMhelix
738 - 760
inside
761 - 812
Population Genetic Test Statistics
Pi
232.533341
Theta
182.568122
Tajima's D
0.630634
CLR
0.303409
CSRT
0.550472476376181
Interpretation
Uncertain
