Gene
KWMTBOMO01375
Pre Gene Modal
BGIBMGA008902
Annotation
PREDICTED:_estradiol_17-beta-dehydrogenase_11-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 2.044
Sequence
CDS
ATGTCTGAACAAACCCAAGAGTATATAAACGGGATGAGACCGGGGCCTCTCACGCGGGAGATCCCGGAGTGTATGAGGGACAAATATACCGTCGTGCAATCAATTATAGACGTTATAGTGTTTGTCATCGTCGGTGTGGGATATGTTATACAAGCGATATACAGGACTATAGTTGGGTATCCTAAGAGAGATCTCAAAGGAGGCGTGGCGGTTGTTACTGGAGGCGGCGGAGGCTTGGGCAGTCTTATAGCCTTGAGGCTAGCGCGACTCGGCTGCACCATTGTCCTGTGGGATATCAATAAGCAAGGTTTGGAAGATACAGTGAAGCTTGTAAAAGGCGTCGGTGGTAAATGCTACGGCTACGTAGTAGATCTCGCCAGCAGGGAAGACATCTACCGTGTCGCTAAAAAAGTTGAAGAGCAGGTTGGGAAGGTAAACCTACTTATCAATAATGCAGGCGTCGTCTCAGGACACTACCTCCTGGAGACTCCAGACCACCTCATTCAAAGGACTTTCGATGTCAACATTCTTGCACATTTCTGGACGGTGAAAGCCTTCCTGCCAACCATGATCAGTCAAAATGATGGTCATATAGTGACCATCGCTTCAATGGCCGGACACGTGGGTGTTGCCAAGCTGGTCGACTACTGTTCGTCAAAAGCTGCTGCCTGCGGCTTCGACGAGGCTCTCAGGGTCGAGCTAGAGACGAAAGGCGTTAAGGGTGTGAAGACTTCGCTGATATGTCCGTACTTCATTCGATCGACGGGCATGTTTGAAGAGGTTAACTCAAGGTTCGTGCCACAGCTGAATTCGAACGAGGTGGCGGATCGTGTTGTGCTCGCGATACGCACCAGTGAGCCTTTCGCTCTAATACCAGGTTTCTTTCGGGTGTTACTACCTTTCAAATGGATAGTACCTTGGCCGTGTATATCAGAGCTGATACGCGGTCTGGTGCCGGACGCGGTCCCTGTGGCCCCTAACGCGCGTATAGAGACACCATCCTCAGCACCAAAAATAGACATGAAAACGGACTCCAGGCCTATGTCACTGCTGCCCCCAGCAAGACACGACCGCCAGGTTTGA
Protein
MSEQTQEYINGMRPGPLTREIPECMRDKYTVVQSIIDVIVFVIVGVGYVIQAIYRTIVGYPKRDLKGGVAVVTGGGGGLGSLIALRLARLGCTIVLWDINKQGLEDTVKLVKGVGGKCYGYVVDLASREDIYRVAKKVEEQVGKVNLLINNAGVVSGHYLLETPDHLIQRTFDVNILAHFWTVKAFLPTMISQNDGHIVTIASMAGHVGVAKLVDYCSSKAAACGFDEALRVELETKGVKGVKTSLICPYFIRSTGMFEEVNSRFVPQLNSNEVADRVVLAIRTSEPFALIPGFFRVLLPFKWIVPWPCISELIRGLVPDAVPVAPNARIETPSSAPKIDMKTDSRPMSLLPPARHDRQV
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
S4PTF3
A0A194RN06
A0A194Q0K1
A0A1Q3G5J9
A0A2A4KBK3
A0A2H1WH70
+ More
A0A1S4FQ43 A0A182QJD4 Q16TB7 A0A182N8S1 A0A182R6X3 A0A182YHE1 A0A1W7R897 U5EXD2 A0A182VSW9 Q7Q0P2 B3MU63 A0A182I0S2 A0A1J1HZH1 B3NKV8 A0A1W7R8Z7 W5JHS0 A0A1Q3G4U4 A0A1Q3G4S7 A0A1S4H3D5 A0A0J9R5G9 B4JEA4 A0A1Q3G4T1 A0A3B0JIE0 T1D655 B4Q3X4 B4IFJ2 B4P708 A0A0J9R697 Q9VIG6 B4MZT9 A0A0M4EC92 T1DGN0 B4LUM3 A0A1W4UVS9 Q8SYX3 Q29NR5 B4GJH1 W8B5P7 T1PKV9 A0A1L8DIT9 A0A1I8MP89 B4KKZ9 A0A1I8PKM0 A0A0A1XI67 A0A034W5U9 A0A0K8WHU1 B0W9U0 A0A1A9WZU0 A0A026VVV7 A0A182XM85 K7ITC0 A0A182P886 A0A088A186
A0A1S4FQ43 A0A182QJD4 Q16TB7 A0A182N8S1 A0A182R6X3 A0A182YHE1 A0A1W7R897 U5EXD2 A0A182VSW9 Q7Q0P2 B3MU63 A0A182I0S2 A0A1J1HZH1 B3NKV8 A0A1W7R8Z7 W5JHS0 A0A1Q3G4U4 A0A1Q3G4S7 A0A1S4H3D5 A0A0J9R5G9 B4JEA4 A0A1Q3G4T1 A0A3B0JIE0 T1D655 B4Q3X4 B4IFJ2 B4P708 A0A0J9R697 Q9VIG6 B4MZT9 A0A0M4EC92 T1DGN0 B4LUM3 A0A1W4UVS9 Q8SYX3 Q29NR5 B4GJH1 W8B5P7 T1PKV9 A0A1L8DIT9 A0A1I8MP89 B4KKZ9 A0A1I8PKM0 A0A0A1XI67 A0A034W5U9 A0A0K8WHU1 B0W9U0 A0A1A9WZU0 A0A026VVV7 A0A182XM85 K7ITC0 A0A182P886 A0A088A186
Pubmed
EMBL
GAIX01012138
JAA80422.1
KQ459989
KPJ18715.1
KQ459591
KPI96940.1
+ More
GEYN01000181 JAV44808.1 NWSH01000002 PCG81052.1 ODYU01008576 SOQ52296.1 AXCN02001862 CH477653 EAT37754.1 GEHC01000256 JAV47389.1 GANO01002593 JAB57278.1 AAAB01008980 EAA13950.4 CH902624 EDV33392.1 APCN01005251 CVRI01000037 CRK93355.1 CH954179 EDV54481.2 GEHC01000035 JAV47610.1 ADMH02001476 ETN62445.1 GFDL01000256 JAV34789.1 GFDL01000235 JAV34810.1 CM002910 KMY91266.1 CH916368 EDW03624.1 GFDL01000241 JAV34804.1 OUUW01000006 SPP82127.1 GALA01000228 JAA94624.1 CM000361 EDX05690.1 CH480833 EDW46414.1 CM000158 EDW89977.1 KMY91264.1 KMY91265.1 AE014134 BT150343 AAF53953.3 AGW30430.1 CH963920 EDW77874.1 CP012523 ALC40351.1 GALA01001586 JAA93266.1 CH940649 EDW64209.1 AY071263 AAL48885.1 CH379058 EAL34579.2 KRT03678.1 KRT03679.1 CH479184 EDW36787.1 GAMC01017944 JAB88611.1 KA649402 AFP64031.1 GFDF01007718 JAV06366.1 CH933807 EDW12749.2 GBXI01003233 JAD11059.1 GAKP01008863 JAC50089.1 GDHF01025215 GDHF01001884 JAI27099.1 JAI50430.1 DS231866 EDS40507.1 KK107796 EZA47616.1 AAZX01003330 AAZX01004515
GEYN01000181 JAV44808.1 NWSH01000002 PCG81052.1 ODYU01008576 SOQ52296.1 AXCN02001862 CH477653 EAT37754.1 GEHC01000256 JAV47389.1 GANO01002593 JAB57278.1 AAAB01008980 EAA13950.4 CH902624 EDV33392.1 APCN01005251 CVRI01000037 CRK93355.1 CH954179 EDV54481.2 GEHC01000035 JAV47610.1 ADMH02001476 ETN62445.1 GFDL01000256 JAV34789.1 GFDL01000235 JAV34810.1 CM002910 KMY91266.1 CH916368 EDW03624.1 GFDL01000241 JAV34804.1 OUUW01000006 SPP82127.1 GALA01000228 JAA94624.1 CM000361 EDX05690.1 CH480833 EDW46414.1 CM000158 EDW89977.1 KMY91264.1 KMY91265.1 AE014134 BT150343 AAF53953.3 AGW30430.1 CH963920 EDW77874.1 CP012523 ALC40351.1 GALA01001586 JAA93266.1 CH940649 EDW64209.1 AY071263 AAL48885.1 CH379058 EAL34579.2 KRT03678.1 KRT03679.1 CH479184 EDW36787.1 GAMC01017944 JAB88611.1 KA649402 AFP64031.1 GFDF01007718 JAV06366.1 CH933807 EDW12749.2 GBXI01003233 JAD11059.1 GAKP01008863 JAC50089.1 GDHF01025215 GDHF01001884 JAI27099.1 JAI50430.1 DS231866 EDS40507.1 KK107796 EZA47616.1 AAZX01003330 AAZX01004515
Proteomes
UP000053240
UP000053268
UP000218220
UP000075886
UP000008820
UP000075884
+ More
UP000075900 UP000076408 UP000075920 UP000007062 UP000007801 UP000075840 UP000183832 UP000008711 UP000000673 UP000001070 UP000268350 UP000000304 UP000001292 UP000002282 UP000000803 UP000007798 UP000092553 UP000008792 UP000192221 UP000001819 UP000008744 UP000095301 UP000009192 UP000095300 UP000002320 UP000091820 UP000053097 UP000076407 UP000002358 UP000075885 UP000005203
UP000075900 UP000076408 UP000075920 UP000007062 UP000007801 UP000075840 UP000183832 UP000008711 UP000000673 UP000001070 UP000268350 UP000000304 UP000001292 UP000002282 UP000000803 UP000007798 UP000092553 UP000008792 UP000192221 UP000001819 UP000008744 UP000095301 UP000009192 UP000095300 UP000002320 UP000091820 UP000053097 UP000076407 UP000002358 UP000075885 UP000005203
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
S4PTF3
A0A194RN06
A0A194Q0K1
A0A1Q3G5J9
A0A2A4KBK3
A0A2H1WH70
+ More
A0A1S4FQ43 A0A182QJD4 Q16TB7 A0A182N8S1 A0A182R6X3 A0A182YHE1 A0A1W7R897 U5EXD2 A0A182VSW9 Q7Q0P2 B3MU63 A0A182I0S2 A0A1J1HZH1 B3NKV8 A0A1W7R8Z7 W5JHS0 A0A1Q3G4U4 A0A1Q3G4S7 A0A1S4H3D5 A0A0J9R5G9 B4JEA4 A0A1Q3G4T1 A0A3B0JIE0 T1D655 B4Q3X4 B4IFJ2 B4P708 A0A0J9R697 Q9VIG6 B4MZT9 A0A0M4EC92 T1DGN0 B4LUM3 A0A1W4UVS9 Q8SYX3 Q29NR5 B4GJH1 W8B5P7 T1PKV9 A0A1L8DIT9 A0A1I8MP89 B4KKZ9 A0A1I8PKM0 A0A0A1XI67 A0A034W5U9 A0A0K8WHU1 B0W9U0 A0A1A9WZU0 A0A026VVV7 A0A182XM85 K7ITC0 A0A182P886 A0A088A186
A0A1S4FQ43 A0A182QJD4 Q16TB7 A0A182N8S1 A0A182R6X3 A0A182YHE1 A0A1W7R897 U5EXD2 A0A182VSW9 Q7Q0P2 B3MU63 A0A182I0S2 A0A1J1HZH1 B3NKV8 A0A1W7R8Z7 W5JHS0 A0A1Q3G4U4 A0A1Q3G4S7 A0A1S4H3D5 A0A0J9R5G9 B4JEA4 A0A1Q3G4T1 A0A3B0JIE0 T1D655 B4Q3X4 B4IFJ2 B4P708 A0A0J9R697 Q9VIG6 B4MZT9 A0A0M4EC92 T1DGN0 B4LUM3 A0A1W4UVS9 Q8SYX3 Q29NR5 B4GJH1 W8B5P7 T1PKV9 A0A1L8DIT9 A0A1I8MP89 B4KKZ9 A0A1I8PKM0 A0A0A1XI67 A0A034W5U9 A0A0K8WHU1 B0W9U0 A0A1A9WZU0 A0A026VVV7 A0A182XM85 K7ITC0 A0A182P886 A0A088A186
PDB
1YB1
E-value=1.07979e-47,
Score=479
Ontologies
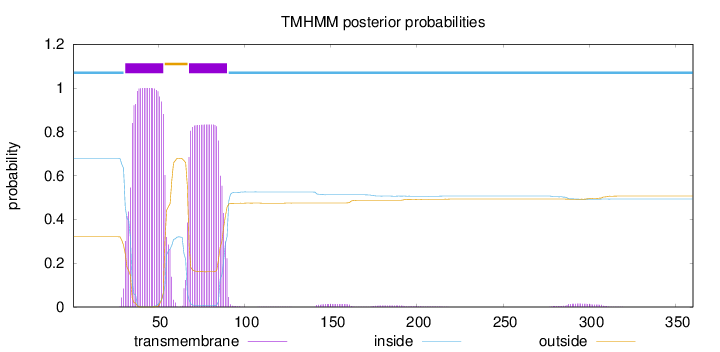
Topology
Length:
360
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.1555400000001
Exp number, first 60 AAs:
21.90877
Total prob of N-in:
0.67945
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 53
outside
54 - 67
TMhelix
68 - 90
inside
91 - 360
Population Genetic Test Statistics
Pi
276.631667
Theta
194.315034
Tajima's D
1.526796
CLR
0.318621
CSRT
0.792610369481526
Interpretation
Uncertain
