Gene
KWMTBOMO01371
Pre Gene Modal
BGIBMGA008987
Annotation
PREDICTED:_epidermal_retinol_dehydrogenase_2_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 2.549
Sequence
CDS
ATGATTAAGAGCGGCAAAGGGCACATAGTCACCGTGGGTTCTGTAGCAGGGCTACTCGGCACGTACCGCTGTACTGATTACAGTGCTACCAAGTTTGCAACAGTGGGCTTCCATGAGAGCCTCTTTACAGAGCTAAGGGCCCACGGTCACAATACAATCCACGCTACCCTTGTATGTCCTTATTATATTAACACGGGAATGTTTAACGGAGTTACTCCTCGTCTGATGCCGATGCTAGAACCGGACTATGTTGCGGAAACAATGATTGACTCCATCAGAAAGAATGAAGTCAATTGCATCATGCCCGGTTCCGTCAGATACTTGTTACCTCTGAAGTGTCTACTCCCAGCTAAAATGTGCTGGGATTTGATGCATCGAGTAATGAAAGGCCCTCAGTCAATGATGGAGTTCAAGGGTAAGCCTAAAGTAGCAGGTGGTTAG
Protein
MIKSGKGHIVTVGSVAGLLGTYRCTDYSATKFATVGFHESLFTELRAHGHNTIHATLVCPYYINTGMFNGVTPRLMPMLEPDYVAETMIDSIRKNEVNCIMPGSVRYLLPLKCLLPAKMCWDLMHRVMKGPQSMMEFKGKPKVAGG
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JHI9
A0A2A4KB77
A0A2W1C020
A0A194RRP2
A0A194PU52
I4DJV0
+ More
A0A212EV09 I4DNX7 A0A2H1V957 Q16PY7 A0A182KB94 A0A182G6H3 A0A084W895 A0A182RVJ6 A0A023EQU1 A0A023EPN3 A0A182PPJ0 A0A023EQU0 A0A182Q8I1 A0A182W730 A0A182T4E5 A0A2M4A3C5 A0A182NUE1 A0A182M205 A0A182Y087 A0A182JEB9 A0A1B0CW19 A0A182TDH4 A0A0L7R5P1 A0A182L1G5 A0A182WU24 Q7Q403 A0A182HUN6 A0A1J1I0Q6 A0A088A1T2 W5J4M7 T1PJH0 A0A0A1X7E9 A0A182UR22 A0A0A1XGY9 A0A1I8NPC2 A0A0N0BCH2 A0A2A3ELT2 A0A0Q9WX42 A0A0L0CPD9 W8C6X1 B4Q328 Q9VR28 A0A1S4KFV9 B4I3C6 A0A0M3QU59 A0A0M4E9N6 B0XIZ4 A0A1W4UVY0 B3ML91 A0A0K8UPA5 B3N498 B4JD24 B4MDE0 B4NZ10 A0A310SLE3 A0A0K8U1U9 A0A1B0G671 A0A336MF17 A0A3B0JUW5 A0A0K8WFD1 A0A336K2S4 Q29MQ7 B4G8X1 A0A1A9UXB0 A0A1A9Y4P0 A0A1B0ATM4 B4N1A9 B4KFR1 A0A1A9ZN68 E0VQE3 K7ITC1 A0A1A9WBJ4 A0A195B671 A0A158NT41 A0A151X0D3 A0A195C3E0 A0A026VV64 A0A154P2Y2 V5FQF8 A0A151JZR9 D6WW87 A0A1W4WER5 A0A0T6AXI0 E2BNA4 A0A3B0K2N7 J3JUE6 U4U187 N6T0W3 E2A0R4 A0A0L7KTJ2 A0A0L8I2T1
A0A212EV09 I4DNX7 A0A2H1V957 Q16PY7 A0A182KB94 A0A182G6H3 A0A084W895 A0A182RVJ6 A0A023EQU1 A0A023EPN3 A0A182PPJ0 A0A023EQU0 A0A182Q8I1 A0A182W730 A0A182T4E5 A0A2M4A3C5 A0A182NUE1 A0A182M205 A0A182Y087 A0A182JEB9 A0A1B0CW19 A0A182TDH4 A0A0L7R5P1 A0A182L1G5 A0A182WU24 Q7Q403 A0A182HUN6 A0A1J1I0Q6 A0A088A1T2 W5J4M7 T1PJH0 A0A0A1X7E9 A0A182UR22 A0A0A1XGY9 A0A1I8NPC2 A0A0N0BCH2 A0A2A3ELT2 A0A0Q9WX42 A0A0L0CPD9 W8C6X1 B4Q328 Q9VR28 A0A1S4KFV9 B4I3C6 A0A0M3QU59 A0A0M4E9N6 B0XIZ4 A0A1W4UVY0 B3ML91 A0A0K8UPA5 B3N498 B4JD24 B4MDE0 B4NZ10 A0A310SLE3 A0A0K8U1U9 A0A1B0G671 A0A336MF17 A0A3B0JUW5 A0A0K8WFD1 A0A336K2S4 Q29MQ7 B4G8X1 A0A1A9UXB0 A0A1A9Y4P0 A0A1B0ATM4 B4N1A9 B4KFR1 A0A1A9ZN68 E0VQE3 K7ITC1 A0A1A9WBJ4 A0A195B671 A0A158NT41 A0A151X0D3 A0A195C3E0 A0A026VV64 A0A154P2Y2 V5FQF8 A0A151JZR9 D6WW87 A0A1W4WER5 A0A0T6AXI0 E2BNA4 A0A3B0K2N7 J3JUE6 U4U187 N6T0W3 E2A0R4 A0A0L7KTJ2 A0A0L8I2T1
Pubmed
19121390
28756777
26354079
22651552
22118469
17510324
+ More
26483478 24438588 24945155 25244985 20966253 12364791 14747013 17210077 20920257 23761445 25315136 25830018 17994087 26108605 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20566863 20075255 21347285 24508170 30249741 18362917 19820115 20798317 22516182 23537049 26227816
26483478 24438588 24945155 25244985 20966253 12364791 14747013 17210077 20920257 23761445 25315136 25830018 17994087 26108605 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20566863 20075255 21347285 24508170 30249741 18362917 19820115 20798317 22516182 23537049 26227816
EMBL
BABH01012598
BABH01012599
BABH01012600
BABH01012601
BABH01012602
NWSH01000002
+ More
PCG81053.1 KZ149894 PZC78857.1 KQ459989 KPJ18716.1 KQ459591 KPI96941.1 AK401568 BAM18190.1 AGBW02012259 OWR45335.1 AK403188 BAM19617.1 ODYU01001339 SOQ37383.1 CH477769 CH477355 EAT36411.1 EAT42804.1 JXUM01045202 JXUM01045203 KQ561430 KXJ78606.1 ATLV01021395 KE525317 KFB46439.1 GAPW01002609 JAC10989.1 GAPW01002610 GAPW01002608 JAC10988.1 GAPW01002607 JAC10991.1 AXCN02000762 GGFK01001966 MBW35287.1 AXCM01001677 AXCM01001678 AJWK01031589 AJWK01031590 KQ414652 KOC66081.1 AAAB01008964 EAA12344.3 APCN01002467 APCN01002468 CVRI01000037 CRK93354.1 ADMH02002111 ETN58876.1 KA648255 AFP62884.1 GBXI01007511 JAD06781.1 GBXI01004097 JAD10195.1 KQ435908 KOX68992.1 KZ288215 PBC32715.1 CH940661 KRF85540.1 JRES01000096 KNC34131.1 GAMC01000882 JAC05674.1 CM000361 CM002910 EDX03744.1 KMY88120.1 AE014134 BT060448 AAF50980.1 ACN23225.1 CH480820 EDW54271.1 CP012523 ALC40093.1 ALC38951.1 DS233415 EDS29876.1 CH902620 EDV31709.1 GDHF01023991 JAI28323.1 CH954177 EDV57768.1 CH916368 EDW04268.1 EDW71201.1 CM000157 EDW87674.1 KQ764881 OAD54369.1 GDHF01031823 JAI20491.1 CCAG010007078 UFQT01000478 SSX24648.1 OUUW01000004 SPP79270.1 GDHF01002739 JAI49575.1 UFQS01000059 UFQT01000059 SSW98767.1 SSX19153.1 CH379060 EAL33636.1 CH479180 EDW28801.1 JXJN01003395 JXJN01003396 CH963920 EDW78119.1 CH933807 EDW11026.1 DS235418 EEB15599.1 AAZX01003330 KQ976585 KYM79755.1 ADTU01002590 KQ982617 KYQ53816.1 KQ978350 KYM94706.1 KK107796 QOIP01000013 EZA47617.1 RLU15222.1 KQ434792 KZC05490.1 GALX01008586 JAB59880.1 KQ981377 KYN42382.1 KQ971361 EFA08169.2 LJIG01022602 KRT79700.1 GL449385 EFN82866.1 SPP79271.1 BT126859 AEE61821.1 KB631786 ERL86068.1 APGK01056809 KB741277 ENN71138.1 GL435626 EFN72967.1 JTDY01005819 KOB66582.1 KQ416705 KOF95654.1
PCG81053.1 KZ149894 PZC78857.1 KQ459989 KPJ18716.1 KQ459591 KPI96941.1 AK401568 BAM18190.1 AGBW02012259 OWR45335.1 AK403188 BAM19617.1 ODYU01001339 SOQ37383.1 CH477769 CH477355 EAT36411.1 EAT42804.1 JXUM01045202 JXUM01045203 KQ561430 KXJ78606.1 ATLV01021395 KE525317 KFB46439.1 GAPW01002609 JAC10989.1 GAPW01002610 GAPW01002608 JAC10988.1 GAPW01002607 JAC10991.1 AXCN02000762 GGFK01001966 MBW35287.1 AXCM01001677 AXCM01001678 AJWK01031589 AJWK01031590 KQ414652 KOC66081.1 AAAB01008964 EAA12344.3 APCN01002467 APCN01002468 CVRI01000037 CRK93354.1 ADMH02002111 ETN58876.1 KA648255 AFP62884.1 GBXI01007511 JAD06781.1 GBXI01004097 JAD10195.1 KQ435908 KOX68992.1 KZ288215 PBC32715.1 CH940661 KRF85540.1 JRES01000096 KNC34131.1 GAMC01000882 JAC05674.1 CM000361 CM002910 EDX03744.1 KMY88120.1 AE014134 BT060448 AAF50980.1 ACN23225.1 CH480820 EDW54271.1 CP012523 ALC40093.1 ALC38951.1 DS233415 EDS29876.1 CH902620 EDV31709.1 GDHF01023991 JAI28323.1 CH954177 EDV57768.1 CH916368 EDW04268.1 EDW71201.1 CM000157 EDW87674.1 KQ764881 OAD54369.1 GDHF01031823 JAI20491.1 CCAG010007078 UFQT01000478 SSX24648.1 OUUW01000004 SPP79270.1 GDHF01002739 JAI49575.1 UFQS01000059 UFQT01000059 SSW98767.1 SSX19153.1 CH379060 EAL33636.1 CH479180 EDW28801.1 JXJN01003395 JXJN01003396 CH963920 EDW78119.1 CH933807 EDW11026.1 DS235418 EEB15599.1 AAZX01003330 KQ976585 KYM79755.1 ADTU01002590 KQ982617 KYQ53816.1 KQ978350 KYM94706.1 KK107796 QOIP01000013 EZA47617.1 RLU15222.1 KQ434792 KZC05490.1 GALX01008586 JAB59880.1 KQ981377 KYN42382.1 KQ971361 EFA08169.2 LJIG01022602 KRT79700.1 GL449385 EFN82866.1 SPP79271.1 BT126859 AEE61821.1 KB631786 ERL86068.1 APGK01056809 KB741277 ENN71138.1 GL435626 EFN72967.1 JTDY01005819 KOB66582.1 KQ416705 KOF95654.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000008820
+ More
UP000075881 UP000069940 UP000249989 UP000030765 UP000075900 UP000075885 UP000075886 UP000075920 UP000075901 UP000075884 UP000075883 UP000076408 UP000075880 UP000092461 UP000075902 UP000053825 UP000075882 UP000076407 UP000007062 UP000075840 UP000183832 UP000005203 UP000000673 UP000095301 UP000075903 UP000095300 UP000053105 UP000242457 UP000008792 UP000037069 UP000000304 UP000000803 UP000001292 UP000092553 UP000002320 UP000192221 UP000007801 UP000008711 UP000001070 UP000002282 UP000092444 UP000268350 UP000001819 UP000008744 UP000078200 UP000092443 UP000092460 UP000007798 UP000009192 UP000092445 UP000009046 UP000002358 UP000091820 UP000078540 UP000005205 UP000075809 UP000078542 UP000053097 UP000279307 UP000076502 UP000078541 UP000007266 UP000192223 UP000008237 UP000030742 UP000019118 UP000000311 UP000037510 UP000053454
UP000075881 UP000069940 UP000249989 UP000030765 UP000075900 UP000075885 UP000075886 UP000075920 UP000075901 UP000075884 UP000075883 UP000076408 UP000075880 UP000092461 UP000075902 UP000053825 UP000075882 UP000076407 UP000007062 UP000075840 UP000183832 UP000005203 UP000000673 UP000095301 UP000075903 UP000095300 UP000053105 UP000242457 UP000008792 UP000037069 UP000000304 UP000000803 UP000001292 UP000092553 UP000002320 UP000192221 UP000007801 UP000008711 UP000001070 UP000002282 UP000092444 UP000268350 UP000001819 UP000008744 UP000078200 UP000092443 UP000092460 UP000007798 UP000009192 UP000092445 UP000009046 UP000002358 UP000091820 UP000078540 UP000005205 UP000075809 UP000078542 UP000053097 UP000279307 UP000076502 UP000078541 UP000007266 UP000192223 UP000008237 UP000030742 UP000019118 UP000000311 UP000037510 UP000053454
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JHI9
A0A2A4KB77
A0A2W1C020
A0A194RRP2
A0A194PU52
I4DJV0
+ More
A0A212EV09 I4DNX7 A0A2H1V957 Q16PY7 A0A182KB94 A0A182G6H3 A0A084W895 A0A182RVJ6 A0A023EQU1 A0A023EPN3 A0A182PPJ0 A0A023EQU0 A0A182Q8I1 A0A182W730 A0A182T4E5 A0A2M4A3C5 A0A182NUE1 A0A182M205 A0A182Y087 A0A182JEB9 A0A1B0CW19 A0A182TDH4 A0A0L7R5P1 A0A182L1G5 A0A182WU24 Q7Q403 A0A182HUN6 A0A1J1I0Q6 A0A088A1T2 W5J4M7 T1PJH0 A0A0A1X7E9 A0A182UR22 A0A0A1XGY9 A0A1I8NPC2 A0A0N0BCH2 A0A2A3ELT2 A0A0Q9WX42 A0A0L0CPD9 W8C6X1 B4Q328 Q9VR28 A0A1S4KFV9 B4I3C6 A0A0M3QU59 A0A0M4E9N6 B0XIZ4 A0A1W4UVY0 B3ML91 A0A0K8UPA5 B3N498 B4JD24 B4MDE0 B4NZ10 A0A310SLE3 A0A0K8U1U9 A0A1B0G671 A0A336MF17 A0A3B0JUW5 A0A0K8WFD1 A0A336K2S4 Q29MQ7 B4G8X1 A0A1A9UXB0 A0A1A9Y4P0 A0A1B0ATM4 B4N1A9 B4KFR1 A0A1A9ZN68 E0VQE3 K7ITC1 A0A1A9WBJ4 A0A195B671 A0A158NT41 A0A151X0D3 A0A195C3E0 A0A026VV64 A0A154P2Y2 V5FQF8 A0A151JZR9 D6WW87 A0A1W4WER5 A0A0T6AXI0 E2BNA4 A0A3B0K2N7 J3JUE6 U4U187 N6T0W3 E2A0R4 A0A0L7KTJ2 A0A0L8I2T1
A0A212EV09 I4DNX7 A0A2H1V957 Q16PY7 A0A182KB94 A0A182G6H3 A0A084W895 A0A182RVJ6 A0A023EQU1 A0A023EPN3 A0A182PPJ0 A0A023EQU0 A0A182Q8I1 A0A182W730 A0A182T4E5 A0A2M4A3C5 A0A182NUE1 A0A182M205 A0A182Y087 A0A182JEB9 A0A1B0CW19 A0A182TDH4 A0A0L7R5P1 A0A182L1G5 A0A182WU24 Q7Q403 A0A182HUN6 A0A1J1I0Q6 A0A088A1T2 W5J4M7 T1PJH0 A0A0A1X7E9 A0A182UR22 A0A0A1XGY9 A0A1I8NPC2 A0A0N0BCH2 A0A2A3ELT2 A0A0Q9WX42 A0A0L0CPD9 W8C6X1 B4Q328 Q9VR28 A0A1S4KFV9 B4I3C6 A0A0M3QU59 A0A0M4E9N6 B0XIZ4 A0A1W4UVY0 B3ML91 A0A0K8UPA5 B3N498 B4JD24 B4MDE0 B4NZ10 A0A310SLE3 A0A0K8U1U9 A0A1B0G671 A0A336MF17 A0A3B0JUW5 A0A0K8WFD1 A0A336K2S4 Q29MQ7 B4G8X1 A0A1A9UXB0 A0A1A9Y4P0 A0A1B0ATM4 B4N1A9 B4KFR1 A0A1A9ZN68 E0VQE3 K7ITC1 A0A1A9WBJ4 A0A195B671 A0A158NT41 A0A151X0D3 A0A195C3E0 A0A026VV64 A0A154P2Y2 V5FQF8 A0A151JZR9 D6WW87 A0A1W4WER5 A0A0T6AXI0 E2BNA4 A0A3B0K2N7 J3JUE6 U4U187 N6T0W3 E2A0R4 A0A0L7KTJ2 A0A0L8I2T1
PDB
1YB1
E-value=1.79301e-15,
Score=195
Ontologies
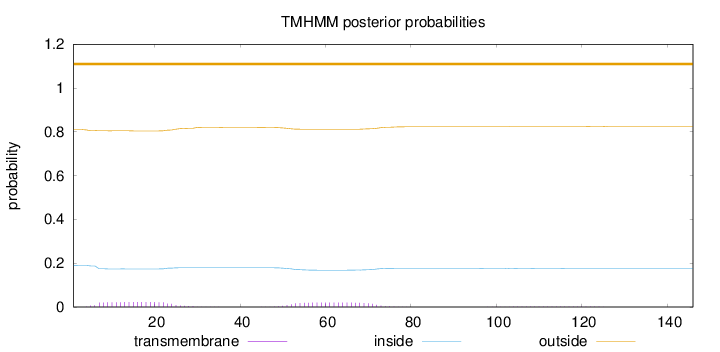
Topology
Length:
146
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.86986
Exp number, first 60 AAs:
0.60243
Total prob of N-in:
0.18991
outside
1 - 146
Population Genetic Test Statistics
Pi
319.467568
Theta
156.258936
Tajima's D
3.076002
CLR
0.160432
CSRT
0.981200939953002
Interpretation
Uncertain
