Gene
KWMTBOMO01369
Pre Gene Modal
BGIBMGA008988
Annotation
PREDICTED:_uncharacterized_protein_LOC101735487_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.901
Sequence
CDS
ATGTGCACTACGAAAGAGTTGAATAAAAGAAGTGCAACGGATGTTAAGTGTCATGAAAACGGAAGGTTTTACAGGAATCCGGATCGAGCTGAAGAAACGGTTTGGACAAGTCAAGAATGCGCGAATTATTACCTTTGCCTGGAAAATGAAGTTTTTGAATTCCATTGCTCGCCGGGACTACTATTCGACGTAAATAGGCAGCTATGTGACAAACAAGAGAATGTACACAACTGCGATCTCACCACAGAGACCAGAGTTCCGAAGCCCCTGCTTGACATGGCTGAGTGTGCGAACGGTACCCATATAGGTTGTTCTGACGGTACCTGTATCCCAGCTGAGTACTTCTGCGATGGATCAATAGACTGTGCGGACTCTTCAGATGAAGGCTGGTGCGATCTAACTTTTGATCCAAATGCTGCAGAACCATGTGACCCTGTCTTTTGCACGTTGCCGGAATGTTTCTGTAGTAAAAATGGTACACAGATACCAGGAAATTTAGTTCCAAGCCAGACTCCACAAATGATTACTCTCACATTTAGCGGAGCTATTAATCATGAAAACTGGGACACGTTTACTAAGCACGTGTTTACTACTGAAAGGAAAAATCCCAACGGGTGTCCATTAAAAGCCACTTTCTTTGTTTCGCACCCATATACGAATTATAGACATGTACAAAAACTTTGGAACGACGGGCATGAAATTGCAGTAAATTCAGTAACGCACCGCGGTCCGGAAGAATGGTGGTCCAAAAACGCGACTGTAGAAGACTGGTTTGACGAAATGGTCGGCGAAGCAAATATTATTAACAGATTCGGTCGTATTCATATGGAAGACTTCAGAGGAATGAGAGTTCCATACCTTTCTGTCGGATGGAATCGTCAATTTCTTATGATGCAAGAATTCGGTTTTGTCTATGACGCTACGATAGTGGCACCGTTAAATGACCCTCCTTTTTGGCCGTATACGCTTGATCATAAAATACCACATTCCTGTACAGGTAAGAATCAATACTGCCCGACAAGAAGTTACGCGAGTCTGTGGGAAATGGTCATAAACCCGCTCGTCAAAGGTGAAAATACATGCGCCATTCTGGATAATTGCCCATTCATTCAAACGGGAGATGACGTTTATGATGCTCTCATAAACAACTTTAAGAGGCATTACCTAACCAATCGTGCACCATTTGGAATTCACTTAAGTTCTACGTGGTTAAGAAATAATGAGTACTTGGTGGCTTTTAAGAATTTCTTGAATGAGTTGCAAAAACTACCTGACGTATACTTTGTCACCTACAAAGAGATACTTGATTGGATAAAGAGACCAACACCGGTGGTACAACTAAAGAAGTTTCAGCCTTGGCAATGTAAGGGGCGACATTTCCGTGAAGCTGAAATAGCCTGTGCCAAACCAAACACATGTAAATTACCTTCCAAAGTATTGGAGCATGATAAATACATGATTACCTGTACTGAATGCCCAAGGAGTTATCCATGGATAAGAAATGAATTCGGATTTGAATAG
Protein
MCTTKELNKRSATDVKCHENGRFYRNPDRAEETVWTSQECANYYLCLENEVFEFHCSPGLLFDVNRQLCDKQENVHNCDLTTETRVPKPLLDMAECANGTHIGCSDGTCIPAEYFCDGSIDCADSSDEGWCDLTFDPNAAEPCDPVFCTLPECFCSKNGTQIPGNLVPSQTPQMITLTFSGAINHENWDTFTKHVFTTERKNPNGCPLKATFFVSHPYTNYRHVQKLWNDGHEIAVNSVTHRGPEEWWSKNATVEDWFDEMVGEANIINRFGRIHMEDFRGMRVPYLSVGWNRQFLMMQEFGFVYDATIVAPLNDPPFWPYTLDHKIPHSCTGKNQYCPTRSYASLWEMVINPLVKGENTCAILDNCPFIQTGDDVYDALINNFKRHYLTNRAPFGIHLSSTWLRNNEYLVAFKNFLNELQKLPDVYFVTYKEILDWIKRPTPVVQLKKFQPWQCKGRHFREAEIACAKPNTCKLPSKVLEHDKYMITCTECPRSYPWIRNEFGFE
Summary
Uniprot
A0A2W1BV56
A0A2H1W5G6
H9JHJ0
A0A2A4KAP7
A0A194PVC5
A0A212EV26
+ More
A0A194RLK6 A0A1S4FBK3 A0A1S4FYE0 A0A182ITH0 Q179B2 A0A182QNG2 B4MYW4 A0A1B0CRU7 Q29MT9 A0A182RVJ4 B4LUF7 A0A182SD46 A0A182Y085 A0A182MA74 B4G8R1 I4DNF0 A0A182WU26 A0A182NUD9 A0A182UUL6 A0A2J7Q955 A0A182HUN8 B4KHX3 A0A1S4GY40 A0A182UJK5 A0A1W4VJG4 A0A182JRT5 A0A336K6E6 B3MK27 B3NNK1 A0A3B0JN38 A0A182F4V5 A0A0K8VLP3 B4I542 A0A0K8V7Y4 Q9VJI8 B4P9A6 B4Q7F7 A0A182PPI8 W8BUP2 B4JBI9 A0A3B0JIP0 A0A182W732 A0A0L0CR97 A0A182H9D2 A0A182GAZ0 Q7Q401 Q16KA8 A0A0A1XEW1 A0A1I8N2V9 A0A084W898 A0A1I8NR28 A0A1I8NR29 A0A1B0AS62 A0A1A9X656 A0A1B0A5W6 W5J651 A0A1J1HZ25 A0A1A9X0P2 D3TL70 A0A2Z4N552 A0A067RVZ5 A0A1A9VTL2 E0VGP9 A0A151JNP9 A0A0L7QV57 A0A151I7D3 A0A151JVE4 E9J1S3 F4WKN7 E2C5L3 A0A195BKV2 E2AJ61 A0A0P5HXL7 A0A0C9QJ10 A0A154PRC6 A0A0P4X5S3 E9G751 A0A1B0FER6 K7ITC3 A0A026WX97 A0A3L8D9H8 A0A1U9XQU5 A0A182L1G7 A0A0P5FFE8 A0A0N8D316 A0A165AII7 A8W488 B0WD94 A0A0K2U2F8 N6TDV2 A0A0N0U745 T1L4A0 A0A1W4WPV9 A0A1W4WDV7
A0A194RLK6 A0A1S4FBK3 A0A1S4FYE0 A0A182ITH0 Q179B2 A0A182QNG2 B4MYW4 A0A1B0CRU7 Q29MT9 A0A182RVJ4 B4LUF7 A0A182SD46 A0A182Y085 A0A182MA74 B4G8R1 I4DNF0 A0A182WU26 A0A182NUD9 A0A182UUL6 A0A2J7Q955 A0A182HUN8 B4KHX3 A0A1S4GY40 A0A182UJK5 A0A1W4VJG4 A0A182JRT5 A0A336K6E6 B3MK27 B3NNK1 A0A3B0JN38 A0A182F4V5 A0A0K8VLP3 B4I542 A0A0K8V7Y4 Q9VJI8 B4P9A6 B4Q7F7 A0A182PPI8 W8BUP2 B4JBI9 A0A3B0JIP0 A0A182W732 A0A0L0CR97 A0A182H9D2 A0A182GAZ0 Q7Q401 Q16KA8 A0A0A1XEW1 A0A1I8N2V9 A0A084W898 A0A1I8NR28 A0A1I8NR29 A0A1B0AS62 A0A1A9X656 A0A1B0A5W6 W5J651 A0A1J1HZ25 A0A1A9X0P2 D3TL70 A0A2Z4N552 A0A067RVZ5 A0A1A9VTL2 E0VGP9 A0A151JNP9 A0A0L7QV57 A0A151I7D3 A0A151JVE4 E9J1S3 F4WKN7 E2C5L3 A0A195BKV2 E2AJ61 A0A0P5HXL7 A0A0C9QJ10 A0A154PRC6 A0A0P4X5S3 E9G751 A0A1B0FER6 K7ITC3 A0A026WX97 A0A3L8D9H8 A0A1U9XQU5 A0A182L1G7 A0A0P5FFE8 A0A0N8D316 A0A165AII7 A8W488 B0WD94 A0A0K2U2F8 N6TDV2 A0A0N0U745 T1L4A0 A0A1W4WPV9 A0A1W4WDV7
Pubmed
28756777
19121390
26354079
22118469
17510324
17994087
+ More
15632085 25244985 22651552 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 24495485 26108605 26483478 25830018 25315136 24438588 20920257 23761445 20353571 24845553 20566863 21282665 21719571 20798317 21292972 20075255 24508170 30249741 20966253 18362917 19820115 23537049
15632085 25244985 22651552 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 24495485 26108605 26483478 25830018 25315136 24438588 20920257 23761445 20353571 24845553 20566863 21282665 21719571 20798317 21292972 20075255 24508170 30249741 20966253 18362917 19820115 23537049
EMBL
KZ149894
PZC78859.1
ODYU01006435
SOQ48277.1
BABH01012603
NWSH01000002
+ More
PCG81056.1 KQ459591 KPI96943.1 AGBW02012259 OWR45333.1 KQ459989 KPJ18718.1 CH477355 EAT42807.1 AXCN02000762 CH963913 EDW77303.1 AJWK01025309 CH379060 EAL33604.1 CH940649 EDW64143.2 AXCM01009005 CH479180 EDW28741.1 AK402877 BAM19440.1 NEVH01016943 PNF25120.1 APCN01002468 CH933807 EDW11255.2 AAAB01008964 UFQS01000065 UFQT01000065 SSW98925.1 SSX19311.1 CH902620 EDV31445.1 CH954179 EDV56652.1 OUUW01000006 SPP82283.1 GDHF01012525 JAI39789.1 CH480822 EDW55498.1 GDHF01017265 JAI35049.1 AE014134 AY118355 AAF53561.2 AAM48384.1 CM000158 EDW90235.2 CM000361 CM002910 EDX05255.1 KMY90564.1 GAMC01003628 JAC02928.1 CH916368 EDW04012.1 SPP82284.1 JRES01000022 KNC34858.1 JXUM01030538 KQ560888 KXJ80470.1 JXUM01051695 KQ561701 KXJ77762.1 EAA12484.3 CH477970 EAT34723.1 GBXI01005264 JAD09028.1 ATLV01021396 KE525317 KFB46442.1 JXJN01002673 JXJN01002674 ADMH02002111 ETN58878.1 CVRI01000037 CRK93352.1 EZ422171 ADD18365.1 MF612052 AWX65384.1 KK852428 KDR24044.1 DS235150 EEB12555.1 KQ978858 KYN27922.1 KQ414727 KOC62542.1 KQ978411 KYM94109.1 KQ981703 KYN37446.1 GL767674 EFZ13264.1 GL888206 EGI65129.1 GL452770 EFN76826.1 KQ976453 KYM85399.1 GL439967 EFN66501.1 GDIQ01226553 GDIQ01210309 GDIQ01102099 GDIQ01102039 JAK25172.1 GBYB01003534 JAG73301.1 KQ435078 KZC14441.1 GDIP01246555 JAI76846.1 GL732534 EFX84707.1 CCAG010014747 AAZX01003330 KK107078 EZA60463.1 QOIP01000011 RLU17137.1 KX427155 AQZ26777.1 GDIQ01258766 JAJ92958.1 GDIP01073719 JAM29996.1 LRGB01000615 KZS17709.1 EU190485 KQ971363 ABW74145.1 EFA07840.1 DS231895 EDS44329.1 HACA01014909 CDW32270.1 APGK01041461 APGK01041462 APGK01041463 APGK01041464 KB740994 ENN75933.1 KQ435713 KOX79308.1 CAEY01001080
PCG81056.1 KQ459591 KPI96943.1 AGBW02012259 OWR45333.1 KQ459989 KPJ18718.1 CH477355 EAT42807.1 AXCN02000762 CH963913 EDW77303.1 AJWK01025309 CH379060 EAL33604.1 CH940649 EDW64143.2 AXCM01009005 CH479180 EDW28741.1 AK402877 BAM19440.1 NEVH01016943 PNF25120.1 APCN01002468 CH933807 EDW11255.2 AAAB01008964 UFQS01000065 UFQT01000065 SSW98925.1 SSX19311.1 CH902620 EDV31445.1 CH954179 EDV56652.1 OUUW01000006 SPP82283.1 GDHF01012525 JAI39789.1 CH480822 EDW55498.1 GDHF01017265 JAI35049.1 AE014134 AY118355 AAF53561.2 AAM48384.1 CM000158 EDW90235.2 CM000361 CM002910 EDX05255.1 KMY90564.1 GAMC01003628 JAC02928.1 CH916368 EDW04012.1 SPP82284.1 JRES01000022 KNC34858.1 JXUM01030538 KQ560888 KXJ80470.1 JXUM01051695 KQ561701 KXJ77762.1 EAA12484.3 CH477970 EAT34723.1 GBXI01005264 JAD09028.1 ATLV01021396 KE525317 KFB46442.1 JXJN01002673 JXJN01002674 ADMH02002111 ETN58878.1 CVRI01000037 CRK93352.1 EZ422171 ADD18365.1 MF612052 AWX65384.1 KK852428 KDR24044.1 DS235150 EEB12555.1 KQ978858 KYN27922.1 KQ414727 KOC62542.1 KQ978411 KYM94109.1 KQ981703 KYN37446.1 GL767674 EFZ13264.1 GL888206 EGI65129.1 GL452770 EFN76826.1 KQ976453 KYM85399.1 GL439967 EFN66501.1 GDIQ01226553 GDIQ01210309 GDIQ01102099 GDIQ01102039 JAK25172.1 GBYB01003534 JAG73301.1 KQ435078 KZC14441.1 GDIP01246555 JAI76846.1 GL732534 EFX84707.1 CCAG010014747 AAZX01003330 KK107078 EZA60463.1 QOIP01000011 RLU17137.1 KX427155 AQZ26777.1 GDIQ01258766 JAJ92958.1 GDIP01073719 JAM29996.1 LRGB01000615 KZS17709.1 EU190485 KQ971363 ABW74145.1 EFA07840.1 DS231895 EDS44329.1 HACA01014909 CDW32270.1 APGK01041461 APGK01041462 APGK01041463 APGK01041464 KB740994 ENN75933.1 KQ435713 KOX79308.1 CAEY01001080
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000075880
+ More
UP000008820 UP000075886 UP000007798 UP000092461 UP000001819 UP000075900 UP000008792 UP000075901 UP000076408 UP000075883 UP000008744 UP000076407 UP000075884 UP000075903 UP000235965 UP000075840 UP000009192 UP000075902 UP000192221 UP000075881 UP000007801 UP000008711 UP000268350 UP000069272 UP000001292 UP000000803 UP000002282 UP000000304 UP000075885 UP000001070 UP000075920 UP000037069 UP000069940 UP000249989 UP000007062 UP000095301 UP000030765 UP000095300 UP000092460 UP000092443 UP000092445 UP000000673 UP000183832 UP000091820 UP000027135 UP000078200 UP000009046 UP000078492 UP000053825 UP000078542 UP000078541 UP000007755 UP000008237 UP000078540 UP000000311 UP000076502 UP000000305 UP000092444 UP000002358 UP000053097 UP000279307 UP000075882 UP000076858 UP000007266 UP000002320 UP000019118 UP000053105 UP000015104 UP000192223
UP000008820 UP000075886 UP000007798 UP000092461 UP000001819 UP000075900 UP000008792 UP000075901 UP000076408 UP000075883 UP000008744 UP000076407 UP000075884 UP000075903 UP000235965 UP000075840 UP000009192 UP000075902 UP000192221 UP000075881 UP000007801 UP000008711 UP000268350 UP000069272 UP000001292 UP000000803 UP000002282 UP000000304 UP000075885 UP000001070 UP000075920 UP000037069 UP000069940 UP000249989 UP000007062 UP000095301 UP000030765 UP000095300 UP000092460 UP000092443 UP000092445 UP000000673 UP000183832 UP000091820 UP000027135 UP000078200 UP000009046 UP000078492 UP000053825 UP000078542 UP000078541 UP000007755 UP000008237 UP000078540 UP000000311 UP000076502 UP000000305 UP000092444 UP000002358 UP000053097 UP000279307 UP000075882 UP000076858 UP000007266 UP000002320 UP000019118 UP000053105 UP000015104 UP000192223
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2W1BV56
A0A2H1W5G6
H9JHJ0
A0A2A4KAP7
A0A194PVC5
A0A212EV26
+ More
A0A194RLK6 A0A1S4FBK3 A0A1S4FYE0 A0A182ITH0 Q179B2 A0A182QNG2 B4MYW4 A0A1B0CRU7 Q29MT9 A0A182RVJ4 B4LUF7 A0A182SD46 A0A182Y085 A0A182MA74 B4G8R1 I4DNF0 A0A182WU26 A0A182NUD9 A0A182UUL6 A0A2J7Q955 A0A182HUN8 B4KHX3 A0A1S4GY40 A0A182UJK5 A0A1W4VJG4 A0A182JRT5 A0A336K6E6 B3MK27 B3NNK1 A0A3B0JN38 A0A182F4V5 A0A0K8VLP3 B4I542 A0A0K8V7Y4 Q9VJI8 B4P9A6 B4Q7F7 A0A182PPI8 W8BUP2 B4JBI9 A0A3B0JIP0 A0A182W732 A0A0L0CR97 A0A182H9D2 A0A182GAZ0 Q7Q401 Q16KA8 A0A0A1XEW1 A0A1I8N2V9 A0A084W898 A0A1I8NR28 A0A1I8NR29 A0A1B0AS62 A0A1A9X656 A0A1B0A5W6 W5J651 A0A1J1HZ25 A0A1A9X0P2 D3TL70 A0A2Z4N552 A0A067RVZ5 A0A1A9VTL2 E0VGP9 A0A151JNP9 A0A0L7QV57 A0A151I7D3 A0A151JVE4 E9J1S3 F4WKN7 E2C5L3 A0A195BKV2 E2AJ61 A0A0P5HXL7 A0A0C9QJ10 A0A154PRC6 A0A0P4X5S3 E9G751 A0A1B0FER6 K7ITC3 A0A026WX97 A0A3L8D9H8 A0A1U9XQU5 A0A182L1G7 A0A0P5FFE8 A0A0N8D316 A0A165AII7 A8W488 B0WD94 A0A0K2U2F8 N6TDV2 A0A0N0U745 T1L4A0 A0A1W4WPV9 A0A1W4WDV7
A0A194RLK6 A0A1S4FBK3 A0A1S4FYE0 A0A182ITH0 Q179B2 A0A182QNG2 B4MYW4 A0A1B0CRU7 Q29MT9 A0A182RVJ4 B4LUF7 A0A182SD46 A0A182Y085 A0A182MA74 B4G8R1 I4DNF0 A0A182WU26 A0A182NUD9 A0A182UUL6 A0A2J7Q955 A0A182HUN8 B4KHX3 A0A1S4GY40 A0A182UJK5 A0A1W4VJG4 A0A182JRT5 A0A336K6E6 B3MK27 B3NNK1 A0A3B0JN38 A0A182F4V5 A0A0K8VLP3 B4I542 A0A0K8V7Y4 Q9VJI8 B4P9A6 B4Q7F7 A0A182PPI8 W8BUP2 B4JBI9 A0A3B0JIP0 A0A182W732 A0A0L0CR97 A0A182H9D2 A0A182GAZ0 Q7Q401 Q16KA8 A0A0A1XEW1 A0A1I8N2V9 A0A084W898 A0A1I8NR28 A0A1I8NR29 A0A1B0AS62 A0A1A9X656 A0A1B0A5W6 W5J651 A0A1J1HZ25 A0A1A9X0P2 D3TL70 A0A2Z4N552 A0A067RVZ5 A0A1A9VTL2 E0VGP9 A0A151JNP9 A0A0L7QV57 A0A151I7D3 A0A151JVE4 E9J1S3 F4WKN7 E2C5L3 A0A195BKV2 E2AJ61 A0A0P5HXL7 A0A0C9QJ10 A0A154PRC6 A0A0P4X5S3 E9G751 A0A1B0FER6 K7ITC3 A0A026WX97 A0A3L8D9H8 A0A1U9XQU5 A0A182L1G7 A0A0P5FFE8 A0A0N8D316 A0A165AII7 A8W488 B0WD94 A0A0K2U2F8 N6TDV2 A0A0N0U745 T1L4A0 A0A1W4WPV9 A0A1W4WDV7
PDB
5ZNT
E-value=1.94691e-95,
Score=892
Ontologies
GO
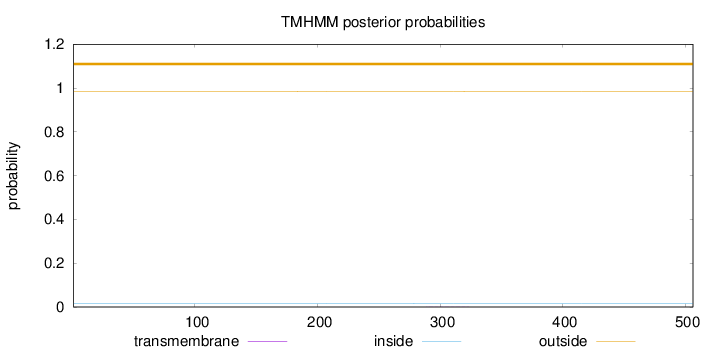
Topology
Length:
506
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01292
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.01524
outside
1 - 506
Population Genetic Test Statistics
Pi
278.282151
Theta
214.855447
Tajima's D
0.929464
CLR
0.215817
CSRT
0.64026798660067
Interpretation
Uncertain
