Gene
KWMTBOMO01365
Pre Gene Modal
BGIBMGA008898
Annotation
PREDICTED:_ATP-dependent_DNA_helicase_PIF1_isoform_X2_[Papilio_xuthus]
Full name
ATP-dependent DNA helicase PIF1
+ More
ATP-dependent DNA helicase
ATP-dependent DNA helicase
Alternative Name
DNA repair and recombination helicase PIF1
Location in the cell
Mitochondrial Reliability : 3.238
Sequence
CDS
ATGTTTTTAGAAGTGACATCCGAAAAACATGCCGCCACTAAATTGGCTCTTAAAAGTTTCAATGTTTTCAAAAAGTTTATGGCTGAAGGTAAAGCAAGTATTAAGTTCCAGGAAGCTAATTGTACAATATTTATTTCAAATGCACCTCCTACAAATTTGGTAATGTTTCTTCGAACTATATTTGTTAAGATGACCAGTGATGAAGAGCAAAAAGCAAATAAAACTCCATCAAAACAGTCAATGAGGGCAAAGCTGTTAAGTGGTAAATCCCAATCATTTGATGAAATAAGTCCAGTTACAGTTGCCGACATTCATATTGCGAAAAGTAAAATTTCAAAATCTACAATCACAACACCATCACCGCCTTCAAAGAAACGAACAGGAAAAAGTTTCTTATTAAAAAGGATTGTGGCAGCTCTACCTCCTGATGTAACGATGCCCACTGCATCCACTGGTGTCGCAGCTTGTCACATTGGTGGTACCACTCTTCATGCTTTTGCTGGCATAGGTGATGGTAGTGGGTCTATCGAAAATTTATGTGAAAGAGCTACTAAAATACCATTGGTAGCACAAAAATGGAGAAAATGTAAACACCTTATTATAGATGAGATATCAATGGTCGATGGAATATTTTTTGAGGTACATAGACAAACAGATCAAGAATTTATTTCGATTCTCAATAGTATCAGAATTGGTCGTGTTACTAAAGAAATCAGCAACAGATTAACAAGAACAGCAAGGCAGAAAATTGAGGCTGATGGCATTTTAGCAACTAGATTATGCTCACATACTAATGACTCAAAAATGATAAATGAGTCAAAATTGATAGCTTTGGATGGAGAGGGAAAATTATTTACATCACAAGACAGTGATAATGCTACTACATTACTAGATATGCAAACTATTGCCCCTTCTAAACTCATGTTAAAAATTGGAGCACAAGTAATGTTATTAAAAAATATTAATGTAAATTCAGGTTTAGTTAATGGAGCCAGAGGAATTGTTGTTCGATTTGAAGAAGGATTCCCTGTTGTTAGATTTAAAAACAAAAAAGAATACATAGCCAGAACAGAACGCTGGTATGTCAAAAATTCGAATGGTACATTATTATGTCGCAGGCAAGTTCCGTTGTCTTTAGCGTGGGCTTTTTCAATACATAAGTCACAAGGATTAACTTTAGACTGTGTTGAGATGTCACTGTCGAAAATATTTGAAGCTGGACAAGCTTATGTTGCATTGAGTAGAGCACAAAGCTTAGACAGCCTTCGTGTATTAGATTTTGATTCCAGACATGTATGGGCAAACCCTGATGTTCTAGAATTTTATCAAAGATTCAGACGAAGATTGCAACAAATGGAGATAGTACCACTTGGTAGACCATTACCTGAAAAAGCAACTAAGAAGTTGAAACTTAGAGATATTTTAGAGAAGCAATTAAGACAGAAATAG
Protein
MFLEVTSEKHAATKLALKSFNVFKKFMAEGKASIKFQEANCTIFISNAPPTNLVMFLRTIFVKMTSDEEQKANKTPSKQSMRAKLLSGKSQSFDEISPVTVADIHIAKSKISKSTITTPSPPSKKRTGKSFLLKRIVAALPPDVTMPTASTGVAACHIGGTTLHAFAGIGDGSGSIENLCERATKIPLVAQKWRKCKHLIIDEISMVDGIFFEVHRQTDQEFISILNSIRIGRVTKEISNRLTRTARQKIEADGILATRLCSHTNDSKMINESKLIALDGEGKLFTSQDSDNATTLLDMQTIAPSKLMLKIGAQVMLLKNINVNSGLVNGARGIVVRFEEGFPVVRFKNKKEYIARTERWYVKNSNGTLLCRRQVPLSLAWAFSIHKSQGLTLDCVEMSLSKIFEAGQAYVALSRAQSLDSLRVLDFDSRHVWANPDVLEFYQRFRRRLQQMEIVPLGRPLPEKATKKLKLRDILEKQLRQK
Summary
Description
DNA-dependent ATPase and 5'-3' DNA helicase required for the maintenance of both mitochondrial and nuclear genome stability.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Subunit
Monomer.
Similarity
Belongs to the helicase family. PIF1 subfamily.
Belongs to the helicase family.
Belongs to the helicase family.
Uniprot
A0A139WDM8
A0A1Y1NF23
A0A182VFF3
J9HF21
A0A195FX53
F4WUX5
+ More
A0A2J7QHS0 A0A232EWR3 A0A158P0R4 A0A0J7KQF7 A0A151WWD1 A0A026WGB1 A0A3L8E2G9 A0A0M9AAC7 A0A151IXN3 T1GYR2 A0A195CXI2 A0A088ASR6 A0A0T6AVX3 E2AZP8 E2C9N0 A0A154PH27 A0A0L7R6X4 A0A1A9ZDD6 A0A1W4XP84 K7J367 A0A2A3E850 A0A182GKW8 A0A1J1I261 A0A2H8TMV8 J9JNH6 A0A0P7YNM5 A0A1A9ZRW9 A0A2S2PYI0 A0A2R7VX24 A0A3M0KBW3 A0A2S2ND12 H0Z9L6 A0A3P8YUH4 A0A3L8SR79 K1PV98 A0A226PIZ2 A0A0Q3RE16 A0A218VFJ6 A0A226MFI1 U3JT03 A0A099ZUG8 A0A3B3ZCX6 A0A210R047 T1J1X4 A0A2P4TED8
A0A2J7QHS0 A0A232EWR3 A0A158P0R4 A0A0J7KQF7 A0A151WWD1 A0A026WGB1 A0A3L8E2G9 A0A0M9AAC7 A0A151IXN3 T1GYR2 A0A195CXI2 A0A088ASR6 A0A0T6AVX3 E2AZP8 E2C9N0 A0A154PH27 A0A0L7R6X4 A0A1A9ZDD6 A0A1W4XP84 K7J367 A0A2A3E850 A0A182GKW8 A0A1J1I261 A0A2H8TMV8 J9JNH6 A0A0P7YNM5 A0A1A9ZRW9 A0A2S2PYI0 A0A2R7VX24 A0A3M0KBW3 A0A2S2ND12 H0Z9L6 A0A3P8YUH4 A0A3L8SR79 K1PV98 A0A226PIZ2 A0A0Q3RE16 A0A218VFJ6 A0A226MFI1 U3JT03 A0A099ZUG8 A0A3B3ZCX6 A0A210R047 T1J1X4 A0A2P4TED8
EC Number
3.6.4.12
Pubmed
EMBL
KQ971361
KYB25955.1
GEZM01003832
JAV96534.1
CH477229
EJY57402.1
+ More
KQ981200 KYN45018.1 GL888378 EGI61989.1 NEVH01013973 PNF28119.1 NNAY01001827 OXU22790.1 ADTU01005769 LBMM01004371 KMQ92494.1 KQ982691 KYQ52047.1 KK107260 EZA54089.1 QOIP01000001 RLU26930.1 KQ435700 KOX80450.1 KQ980819 KYN12681.1 CAQQ02375316 KQ977141 KYN05356.1 LJIG01022674 KRT79311.1 GL444275 EFN61107.1 GL453868 EFN75369.1 KQ434890 KZC10480.1 KQ414646 KOC66579.1 KZ288334 PBC27878.1 JXUM01070933 KQ562635 KXJ75449.1 CVRI01000037 CRK93676.1 GFXV01003515 MBW15320.1 ABLF02025680 JARO02003963 KPP69424.1 GGMS01001276 MBY70479.1 KK854143 PTY12057.1 QRBI01000117 RMC08560.1 GGMR01002389 MBY15008.1 ABQF01041509 ABQF01041510 ABQF01041511 QUSF01000010 RLW06011.1 JH816145 EKC25648.1 AWGT02000057 OXB80076.1 LMAW01001336 KQK83793.1 MUZQ01000002 OWK64451.1 MCFN01000997 OXB54052.1 AGTO01010966 KL898352 KGL85436.1 NEDP02001046 OWF54393.1 JH431790 PPHD01001327 POI34740.1
KQ981200 KYN45018.1 GL888378 EGI61989.1 NEVH01013973 PNF28119.1 NNAY01001827 OXU22790.1 ADTU01005769 LBMM01004371 KMQ92494.1 KQ982691 KYQ52047.1 KK107260 EZA54089.1 QOIP01000001 RLU26930.1 KQ435700 KOX80450.1 KQ980819 KYN12681.1 CAQQ02375316 KQ977141 KYN05356.1 LJIG01022674 KRT79311.1 GL444275 EFN61107.1 GL453868 EFN75369.1 KQ434890 KZC10480.1 KQ414646 KOC66579.1 KZ288334 PBC27878.1 JXUM01070933 KQ562635 KXJ75449.1 CVRI01000037 CRK93676.1 GFXV01003515 MBW15320.1 ABLF02025680 JARO02003963 KPP69424.1 GGMS01001276 MBY70479.1 KK854143 PTY12057.1 QRBI01000117 RMC08560.1 GGMR01002389 MBY15008.1 ABQF01041509 ABQF01041510 ABQF01041511 QUSF01000010 RLW06011.1 JH816145 EKC25648.1 AWGT02000057 OXB80076.1 LMAW01001336 KQK83793.1 MUZQ01000002 OWK64451.1 MCFN01000997 OXB54052.1 AGTO01010966 KL898352 KGL85436.1 NEDP02001046 OWF54393.1 JH431790 PPHD01001327 POI34740.1
Proteomes
UP000007266
UP000075903
UP000008820
UP000078541
UP000007755
UP000235965
+ More
UP000215335 UP000005205 UP000036403 UP000075809 UP000053097 UP000279307 UP000053105 UP000078492 UP000015102 UP000078542 UP000005203 UP000000311 UP000008237 UP000076502 UP000053825 UP000092445 UP000192223 UP000002358 UP000242457 UP000069940 UP000249989 UP000183832 UP000007819 UP000034805 UP000269221 UP000007754 UP000265140 UP000276834 UP000005408 UP000198419 UP000051836 UP000197619 UP000198323 UP000016665 UP000053641 UP000261520 UP000242188
UP000215335 UP000005205 UP000036403 UP000075809 UP000053097 UP000279307 UP000053105 UP000078492 UP000015102 UP000078542 UP000005203 UP000000311 UP000008237 UP000076502 UP000053825 UP000092445 UP000192223 UP000002358 UP000242457 UP000069940 UP000249989 UP000183832 UP000007819 UP000034805 UP000269221 UP000007754 UP000265140 UP000276834 UP000005408 UP000198419 UP000051836 UP000197619 UP000198323 UP000016665 UP000053641 UP000261520 UP000242188
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A139WDM8
A0A1Y1NF23
A0A182VFF3
J9HF21
A0A195FX53
F4WUX5
+ More
A0A2J7QHS0 A0A232EWR3 A0A158P0R4 A0A0J7KQF7 A0A151WWD1 A0A026WGB1 A0A3L8E2G9 A0A0M9AAC7 A0A151IXN3 T1GYR2 A0A195CXI2 A0A088ASR6 A0A0T6AVX3 E2AZP8 E2C9N0 A0A154PH27 A0A0L7R6X4 A0A1A9ZDD6 A0A1W4XP84 K7J367 A0A2A3E850 A0A182GKW8 A0A1J1I261 A0A2H8TMV8 J9JNH6 A0A0P7YNM5 A0A1A9ZRW9 A0A2S2PYI0 A0A2R7VX24 A0A3M0KBW3 A0A2S2ND12 H0Z9L6 A0A3P8YUH4 A0A3L8SR79 K1PV98 A0A226PIZ2 A0A0Q3RE16 A0A218VFJ6 A0A226MFI1 U3JT03 A0A099ZUG8 A0A3B3ZCX6 A0A210R047 T1J1X4 A0A2P4TED8
A0A2J7QHS0 A0A232EWR3 A0A158P0R4 A0A0J7KQF7 A0A151WWD1 A0A026WGB1 A0A3L8E2G9 A0A0M9AAC7 A0A151IXN3 T1GYR2 A0A195CXI2 A0A088ASR6 A0A0T6AVX3 E2AZP8 E2C9N0 A0A154PH27 A0A0L7R6X4 A0A1A9ZDD6 A0A1W4XP84 K7J367 A0A2A3E850 A0A182GKW8 A0A1J1I261 A0A2H8TMV8 J9JNH6 A0A0P7YNM5 A0A1A9ZRW9 A0A2S2PYI0 A0A2R7VX24 A0A3M0KBW3 A0A2S2ND12 H0Z9L6 A0A3P8YUH4 A0A3L8SR79 K1PV98 A0A226PIZ2 A0A0Q3RE16 A0A218VFJ6 A0A226MFI1 U3JT03 A0A099ZUG8 A0A3B3ZCX6 A0A210R047 T1J1X4 A0A2P4TED8
PDB
5FHH
E-value=3.21987e-76,
Score=726
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Mitochondrion
Mitochondrion
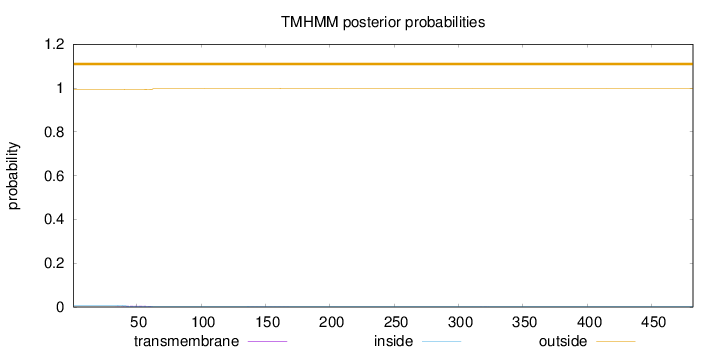
Length:
482
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11683
Exp number, first 60 AAs:
0.09751
Total prob of N-in:
0.00736
outside
1 - 482
Population Genetic Test Statistics
Pi
253.837075
Theta
199.703053
Tajima's D
0.848831
CLR
0
CSRT
0.613769311534423
Interpretation
Uncertain
