Gene
KWMTBOMO01359
Pre Gene Modal
BGIBMGA008896
Annotation
PREDICTED:_SPRY_domain-containing_SOCS_box_protein_4_isoform_X3_[Bombyx_mori]
Full name
Protein gustavus
Location in the cell
Mitochondrial Reliability : 2.759
Sequence
CDS
ATGAAATTTTCCAGGTCATTAAACATATTCGTTAAGGAAGAAGACGCACTGACATTCCACCGGCACCCGGTCGCGCAGAGCACGGACTGCATCCGCGGCCGCACGGGCTACACGCGCGGGCTGCACTGCTGGGAGGTGGTGTGGCCGGCACGACAGCGCGGCACGCACGCCGTCGTGGGCGTCGCCACGTCCCACGCGCCCCTGCACTCAGTCGGCTATCAGAGTCTAGTCGGGGCCACCGATCAGAGCTGGGGCTGGGATTTGGGTAGAAATAAGGTGTTCCATAACGCGAAAGGCACGGGCGGCGGCGGCACGACGTACCCGGCGCTGCTGCGGCCCGACGAACAGTTCCTCGTGCCGGACCGCGTGCTCGTCGTGCTCGACATGGACGAGGGCACGCTCGCCTTCTGCGCCGACGGACGCTACCTCGGCGTCGCGGCGCGCGGCCTGCGCGGCAAGACGCTCTACCCCATCGTCTCCGCCGTCTGGGGGCACGCCGAGATCACCATGAAGTACATCGGCGGGCTAGATCCGGAGCCGCTGCCGCTGATGGAGCTGTGCCGGCGCGTGATCCGGCAGCGCGTGGGCCGGGGCCGGCTGCGCGCGGCGGCGTCCCGCCTGTCGCTCCCGCCCGCGCTCACGGCGTACCTGCTGTACCGCGCGCCCTAG
Protein
MKFSRSLNIFVKEEDALTFHRHPVAQSTDCIRGRTGYTRGLHCWEVVWPARQRGTHAVVGVATSHAPLHSVGYQSLVGATDQSWGWDLGRNKVFHNAKGTGGGGTTYPALLRPDEQFLVPDRVLVVLDMDEGTLAFCADGRYLGVAARGLRGKTLYPIVSAVWGHAEITMKYIGGLDPEPLPLMELCRRVIRQRVGRGRLRAAASRLSLPPALTAYLLYRAP
Summary
Description
Involved in the localization of vas to the posterior pole of the oocyte. Required maternally in the germ line for efficient primordial germ cell formation.
Subunit
Interacts (via B30.2/SPRY domain) with vas; this interaction may be necessary for the transport of vas to the posterior pole of the oocyte. Interacts with Cul-5. May associate with the Elongin BC complex composed of Elongin-B and Elongin-C.
Similarity
Belongs to the SPSB family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Developmental protein
Nucleus
Reference proteome
Feature
chain Protein gustavus
Uniprot
A0A194PU64
A0A194RMH5
A0A2W1BUZ0
A0A2A4KA21
A0A2H1VDP2
A0A2A4KAG1
+ More
A0A2A4KBM1 A0A212EV05 H9JH99 A0A1B0DDB7 A0A0L7LPW5 A0A1L8E317 A0A1L8E3C1 A0A2S2P9H0 A0A336JXF6 V5GX37 A0A2S2Q5E9 A0A1B6CMD4 A0A1Y1KHX1 J9JME6 A0A0T6AVZ6 A0A023EQG6 N6TIS0 A0A182G3Y2 U4U4N0 A0A0A9WYG3 Q16R56 A0A1S4FSF0 A0A1B6JLE3 A0A1Q3G086 A0A1Q3EVH6 A0A1S4JHL1 B0WF17 A0A1B6GY30 A0A1B6MMS3 A0A026WPI5 A0A2R7WZT5 A0A1B6DY31 A0A0K8S3Q8 A0A1W4VWB8 A0A1B6DJZ2 A0A1B6CWC1 E0VX79 A0A139WD54 A0A2P8ZPG3 A0A0N8ECC1 V9IE16 A0A088AVN3 A0A0B4K6X3 A1Z6E0 A0A1I8MGU1 A0A0V0GBT4 A0A023F5R1 A0A069DRM1 A0A0Q9XHV8 A0A0K8S3S0 B4KJG5 A0A0P4VLA9 E2AGK4 A0A0J9R7C8 U5EZK6 A0A1W4X9I2 A0A0L7RIM2 B4IKU4 B4R4G4 A0A0J9R660 A0A0R1EEH4 A0A0Q5WP41 B4ISV9 A0A0N0U603 B4LR16 B5DSW6 B4HCC7 B3N433 A0A146M7L7 A0A0Q9WKT9 W8C9Z7 A0A0K8URB7 B4JP33 W8BZP6 A0A0J9R5X0 A0A1W4XK03 A0A1Y9G6S8 A0A3Q0IR59 A0A182FMT0 A0A1I8PTM8 A0A2M4AR66 A0A1B6MIV9 A0A182IQD7 A0A0A1WIE1 A0A0M4EQW5 A0A0A1WNH1 B4MUD2 E9GL80 W5JTT4 A0A195CNG9 A0A2M3ZJI0 A0A2M4CFT0 A0A0Q9WQH4 A0A2M4BSK2
A0A2A4KBM1 A0A212EV05 H9JH99 A0A1B0DDB7 A0A0L7LPW5 A0A1L8E317 A0A1L8E3C1 A0A2S2P9H0 A0A336JXF6 V5GX37 A0A2S2Q5E9 A0A1B6CMD4 A0A1Y1KHX1 J9JME6 A0A0T6AVZ6 A0A023EQG6 N6TIS0 A0A182G3Y2 U4U4N0 A0A0A9WYG3 Q16R56 A0A1S4FSF0 A0A1B6JLE3 A0A1Q3G086 A0A1Q3EVH6 A0A1S4JHL1 B0WF17 A0A1B6GY30 A0A1B6MMS3 A0A026WPI5 A0A2R7WZT5 A0A1B6DY31 A0A0K8S3Q8 A0A1W4VWB8 A0A1B6DJZ2 A0A1B6CWC1 E0VX79 A0A139WD54 A0A2P8ZPG3 A0A0N8ECC1 V9IE16 A0A088AVN3 A0A0B4K6X3 A1Z6E0 A0A1I8MGU1 A0A0V0GBT4 A0A023F5R1 A0A069DRM1 A0A0Q9XHV8 A0A0K8S3S0 B4KJG5 A0A0P4VLA9 E2AGK4 A0A0J9R7C8 U5EZK6 A0A1W4X9I2 A0A0L7RIM2 B4IKU4 B4R4G4 A0A0J9R660 A0A0R1EEH4 A0A0Q5WP41 B4ISV9 A0A0N0U603 B4LR16 B5DSW6 B4HCC7 B3N433 A0A146M7L7 A0A0Q9WKT9 W8C9Z7 A0A0K8URB7 B4JP33 W8BZP6 A0A0J9R5X0 A0A1W4XK03 A0A1Y9G6S8 A0A3Q0IR59 A0A182FMT0 A0A1I8PTM8 A0A2M4AR66 A0A1B6MIV9 A0A182IQD7 A0A0A1WIE1 A0A0M4EQW5 A0A0A1WNH1 B4MUD2 E9GL80 W5JTT4 A0A195CNG9 A0A2M3ZJI0 A0A2M4CFT0 A0A0Q9WQH4 A0A2M4BSK2
Pubmed
26354079
28756777
22118469
19121390
26227816
28004739
+ More
24945155 23537049 26483478 25401762 17510324 24508170 20566863 18362917 19820115 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 12479811 20123973 16498413 17189197 25315136 25474469 26334808 17994087 27129103 20798317 22936249 17550304 15632085 26823975 24495485 25830018 21292972 20920257 23761445
24945155 23537049 26483478 25401762 17510324 24508170 20566863 18362917 19820115 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 12479811 20123973 16498413 17189197 25315136 25474469 26334808 17994087 27129103 20798317 22936249 17550304 15632085 26823975 24495485 25830018 21292972 20920257 23761445
EMBL
KQ459591
KPI96951.1
KQ459989
KPJ18727.1
KZ149894
PZC78868.1
+ More
NWSH01000002 PCG81071.1 ODYU01001751 SOQ38384.1 PCG81069.1 PCG81072.1 AGBW02012259 OWR45322.1 BABH01012612 AJVK01005388 AJVK01005389 JTDY01000427 KOB77226.1 GFDF01000951 JAV13133.1 GFDF01000950 JAV13134.1 GGMR01013423 MBY26042.1 UFQS01000004 UFQT01000004 SSW96807.1 SSX17194.1 GALX01002134 JAB66332.1 GGMS01003782 MBY72985.1 GEDC01029542 GEDC01022706 GEDC01019871 GEDC01006089 GEDC01004115 GEDC01000749 JAS07756.1 JAS14592.1 JAS17427.1 JAS31209.1 JAS33183.1 JAS36549.1 GEZM01083737 GEZM01083736 JAV61053.1 ABLF02039268 LJIG01022692 KRT79217.1 GAPW01002255 JAC11343.1 APGK01036343 KB740939 ENN77728.1 JXUM01006998 KQ560228 KXJ83733.1 KB632013 ERL88022.1 GBHO01032056 JAG11548.1 CH477724 EAT36876.1 EJY57897.1 GECU01008003 JAS99703.1 GFDL01001845 JAV33200.1 GFDL01015738 JAV19307.1 DS231913 EDS25925.1 GECZ01002440 JAS67329.1 GEBQ01027063 GEBQ01003031 GEBQ01002812 JAT12914.1 JAT36946.1 JAT37165.1 KK107138 EZA57967.1 KK856155 PTY25047.1 GEDC01027969 GEDC01006712 JAS09329.1 JAS30586.1 GBRD01017900 GBRD01017899 GBRD01016176 JAG47927.1 GEDC01024044 GEDC01022326 GEDC01021854 GEDC01017080 GEDC01011323 GEDC01000250 JAS13254.1 JAS14972.1 JAS15444.1 JAS20218.1 JAS25975.1 JAS37048.1 GEDC01019496 GEDC01019159 JAS17802.1 JAS18139.1 DS235829 EEB17985.1 KQ971361 KYB25844.1 PYGN01000003 PSN58391.1 GDIQ01040700 JAN54037.1 JR040932 AEY59330.1 AE013599 AFH07895.1 AY119609 BT089043 BT099916 BT126146 AAM50263.1 AAM68373.2 ADZ74173.1 GECL01000618 JAP05506.1 GBBI01002204 JAC16508.1 GBGD01002171 JAC86718.1 CH933807 KRG03857.1 GBRD01017896 JAG47931.1 EDW13545.1 GDKW01003191 JAI53404.1 GL439322 EFN67424.1 CM002911 KMY91599.1 KMY91601.1 GANO01000623 JAB59248.1 KQ414582 KOC70812.1 CH480857 EDW52702.1 CM000366 EDX17054.1 KMY91596.1 KMY91598.1 KMY91600.1 CH891608 KRK05127.1 CH954177 KQS71058.1 EDW99541.1 KQ435745 KOX76621.1 CH940649 EDW64555.1 CH475636 EDY70908.1 CH479304 EDW26091.1 EDV59927.1 GDHC01003354 JAQ15275.1 KRF81716.1 GAMC01001714 JAC04842.1 GDHF01023251 JAI29063.1 CH916372 EDV99458.1 GAMC01001713 JAC04843.1 KMY91597.1 GGFK01009948 MBW43269.1 GEBQ01004134 JAT35843.1 GBXI01016022 JAC98269.1 CP012523 ALC39195.1 GBXI01014071 GBXI01006904 JAD00221.1 JAD07388.1 CH963857 EDW76058.1 GL732550 EFX79840.1 ADMH02000157 ETN67541.1 KQ977595 KYN01634.1 GGFM01007847 MBW28598.1 GGFL01000018 MBW64196.1 KRF98369.1 GGFJ01006925 MBW56066.1
NWSH01000002 PCG81071.1 ODYU01001751 SOQ38384.1 PCG81069.1 PCG81072.1 AGBW02012259 OWR45322.1 BABH01012612 AJVK01005388 AJVK01005389 JTDY01000427 KOB77226.1 GFDF01000951 JAV13133.1 GFDF01000950 JAV13134.1 GGMR01013423 MBY26042.1 UFQS01000004 UFQT01000004 SSW96807.1 SSX17194.1 GALX01002134 JAB66332.1 GGMS01003782 MBY72985.1 GEDC01029542 GEDC01022706 GEDC01019871 GEDC01006089 GEDC01004115 GEDC01000749 JAS07756.1 JAS14592.1 JAS17427.1 JAS31209.1 JAS33183.1 JAS36549.1 GEZM01083737 GEZM01083736 JAV61053.1 ABLF02039268 LJIG01022692 KRT79217.1 GAPW01002255 JAC11343.1 APGK01036343 KB740939 ENN77728.1 JXUM01006998 KQ560228 KXJ83733.1 KB632013 ERL88022.1 GBHO01032056 JAG11548.1 CH477724 EAT36876.1 EJY57897.1 GECU01008003 JAS99703.1 GFDL01001845 JAV33200.1 GFDL01015738 JAV19307.1 DS231913 EDS25925.1 GECZ01002440 JAS67329.1 GEBQ01027063 GEBQ01003031 GEBQ01002812 JAT12914.1 JAT36946.1 JAT37165.1 KK107138 EZA57967.1 KK856155 PTY25047.1 GEDC01027969 GEDC01006712 JAS09329.1 JAS30586.1 GBRD01017900 GBRD01017899 GBRD01016176 JAG47927.1 GEDC01024044 GEDC01022326 GEDC01021854 GEDC01017080 GEDC01011323 GEDC01000250 JAS13254.1 JAS14972.1 JAS15444.1 JAS20218.1 JAS25975.1 JAS37048.1 GEDC01019496 GEDC01019159 JAS17802.1 JAS18139.1 DS235829 EEB17985.1 KQ971361 KYB25844.1 PYGN01000003 PSN58391.1 GDIQ01040700 JAN54037.1 JR040932 AEY59330.1 AE013599 AFH07895.1 AY119609 BT089043 BT099916 BT126146 AAM50263.1 AAM68373.2 ADZ74173.1 GECL01000618 JAP05506.1 GBBI01002204 JAC16508.1 GBGD01002171 JAC86718.1 CH933807 KRG03857.1 GBRD01017896 JAG47931.1 EDW13545.1 GDKW01003191 JAI53404.1 GL439322 EFN67424.1 CM002911 KMY91599.1 KMY91601.1 GANO01000623 JAB59248.1 KQ414582 KOC70812.1 CH480857 EDW52702.1 CM000366 EDX17054.1 KMY91596.1 KMY91598.1 KMY91600.1 CH891608 KRK05127.1 CH954177 KQS71058.1 EDW99541.1 KQ435745 KOX76621.1 CH940649 EDW64555.1 CH475636 EDY70908.1 CH479304 EDW26091.1 EDV59927.1 GDHC01003354 JAQ15275.1 KRF81716.1 GAMC01001714 JAC04842.1 GDHF01023251 JAI29063.1 CH916372 EDV99458.1 GAMC01001713 JAC04843.1 KMY91597.1 GGFK01009948 MBW43269.1 GEBQ01004134 JAT35843.1 GBXI01016022 JAC98269.1 CP012523 ALC39195.1 GBXI01014071 GBXI01006904 JAD00221.1 JAD07388.1 CH963857 EDW76058.1 GL732550 EFX79840.1 ADMH02000157 ETN67541.1 KQ977595 KYN01634.1 GGFM01007847 MBW28598.1 GGFL01000018 MBW64196.1 KRF98369.1 GGFJ01006925 MBW56066.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000005204
UP000092462
+ More
UP000037510 UP000007819 UP000019118 UP000069940 UP000249989 UP000030742 UP000008820 UP000002320 UP000053097 UP000192221 UP000009046 UP000007266 UP000245037 UP000005203 UP000000803 UP000095301 UP000009192 UP000000311 UP000192223 UP000053825 UP000001292 UP000000304 UP000002282 UP000008711 UP000053105 UP000008792 UP000001819 UP000008744 UP000001070 UP000069272 UP000079169 UP000095300 UP000075880 UP000092553 UP000007798 UP000000305 UP000000673 UP000078542
UP000037510 UP000007819 UP000019118 UP000069940 UP000249989 UP000030742 UP000008820 UP000002320 UP000053097 UP000192221 UP000009046 UP000007266 UP000245037 UP000005203 UP000000803 UP000095301 UP000009192 UP000000311 UP000192223 UP000053825 UP000001292 UP000000304 UP000002282 UP000008711 UP000053105 UP000008792 UP000001819 UP000008744 UP000001070 UP000069272 UP000079169 UP000095300 UP000075880 UP000092553 UP000007798 UP000000305 UP000000673 UP000078542
Interpro
ProteinModelPortal
A0A194PU64
A0A194RMH5
A0A2W1BUZ0
A0A2A4KA21
A0A2H1VDP2
A0A2A4KAG1
+ More
A0A2A4KBM1 A0A212EV05 H9JH99 A0A1B0DDB7 A0A0L7LPW5 A0A1L8E317 A0A1L8E3C1 A0A2S2P9H0 A0A336JXF6 V5GX37 A0A2S2Q5E9 A0A1B6CMD4 A0A1Y1KHX1 J9JME6 A0A0T6AVZ6 A0A023EQG6 N6TIS0 A0A182G3Y2 U4U4N0 A0A0A9WYG3 Q16R56 A0A1S4FSF0 A0A1B6JLE3 A0A1Q3G086 A0A1Q3EVH6 A0A1S4JHL1 B0WF17 A0A1B6GY30 A0A1B6MMS3 A0A026WPI5 A0A2R7WZT5 A0A1B6DY31 A0A0K8S3Q8 A0A1W4VWB8 A0A1B6DJZ2 A0A1B6CWC1 E0VX79 A0A139WD54 A0A2P8ZPG3 A0A0N8ECC1 V9IE16 A0A088AVN3 A0A0B4K6X3 A1Z6E0 A0A1I8MGU1 A0A0V0GBT4 A0A023F5R1 A0A069DRM1 A0A0Q9XHV8 A0A0K8S3S0 B4KJG5 A0A0P4VLA9 E2AGK4 A0A0J9R7C8 U5EZK6 A0A1W4X9I2 A0A0L7RIM2 B4IKU4 B4R4G4 A0A0J9R660 A0A0R1EEH4 A0A0Q5WP41 B4ISV9 A0A0N0U603 B4LR16 B5DSW6 B4HCC7 B3N433 A0A146M7L7 A0A0Q9WKT9 W8C9Z7 A0A0K8URB7 B4JP33 W8BZP6 A0A0J9R5X0 A0A1W4XK03 A0A1Y9G6S8 A0A3Q0IR59 A0A182FMT0 A0A1I8PTM8 A0A2M4AR66 A0A1B6MIV9 A0A182IQD7 A0A0A1WIE1 A0A0M4EQW5 A0A0A1WNH1 B4MUD2 E9GL80 W5JTT4 A0A195CNG9 A0A2M3ZJI0 A0A2M4CFT0 A0A0Q9WQH4 A0A2M4BSK2
A0A2A4KBM1 A0A212EV05 H9JH99 A0A1B0DDB7 A0A0L7LPW5 A0A1L8E317 A0A1L8E3C1 A0A2S2P9H0 A0A336JXF6 V5GX37 A0A2S2Q5E9 A0A1B6CMD4 A0A1Y1KHX1 J9JME6 A0A0T6AVZ6 A0A023EQG6 N6TIS0 A0A182G3Y2 U4U4N0 A0A0A9WYG3 Q16R56 A0A1S4FSF0 A0A1B6JLE3 A0A1Q3G086 A0A1Q3EVH6 A0A1S4JHL1 B0WF17 A0A1B6GY30 A0A1B6MMS3 A0A026WPI5 A0A2R7WZT5 A0A1B6DY31 A0A0K8S3Q8 A0A1W4VWB8 A0A1B6DJZ2 A0A1B6CWC1 E0VX79 A0A139WD54 A0A2P8ZPG3 A0A0N8ECC1 V9IE16 A0A088AVN3 A0A0B4K6X3 A1Z6E0 A0A1I8MGU1 A0A0V0GBT4 A0A023F5R1 A0A069DRM1 A0A0Q9XHV8 A0A0K8S3S0 B4KJG5 A0A0P4VLA9 E2AGK4 A0A0J9R7C8 U5EZK6 A0A1W4X9I2 A0A0L7RIM2 B4IKU4 B4R4G4 A0A0J9R660 A0A0R1EEH4 A0A0Q5WP41 B4ISV9 A0A0N0U603 B4LR16 B5DSW6 B4HCC7 B3N433 A0A146M7L7 A0A0Q9WKT9 W8C9Z7 A0A0K8URB7 B4JP33 W8BZP6 A0A0J9R5X0 A0A1W4XK03 A0A1Y9G6S8 A0A3Q0IR59 A0A182FMT0 A0A1I8PTM8 A0A2M4AR66 A0A1B6MIV9 A0A182IQD7 A0A0A1WIE1 A0A0M4EQW5 A0A0A1WNH1 B4MUD2 E9GL80 W5JTT4 A0A195CNG9 A0A2M3ZJI0 A0A2M4CFT0 A0A0Q9WQH4 A0A2M4BSK2
PDB
2FNJ
E-value=7.31927e-85,
Score=797
Ontologies
GO
GO:0035556
GO:0005737
GO:0007314
GO:0005634
GO:0007280
GO:0005938
GO:0008104
GO:0045495
GO:0035017
GO:0007281
GO:0070449
GO:0031466
GO:0007315
GO:0045732
GO:0046843
GO:0007472
GO:0048471
GO:0016021
GO:0043161
GO:0019005
GO:0006511
GO:0005515
GO:0003676
GO:0005524
GO:0008026
GO:0016820
GO:0016042
GO:0006334
GO:0003779
Topology
Subcellular location
Cytoplasm
Component of the cytoplasmic ribonucleoprotein (RNP) bodies which concentrate at the perinuclear region of the nurse cell and in punctate aggregates throughout the cytoplasm. Some cortical enrichment of these aggregates is also observed. At stage 7, accumulates in the anterior cytoplasm of the oocyte, whereas during stage 10 accumulates at the posterior pole. This posterior distribution is not maintained in later developmental stages. With evidence from 4 publications.
Nucleus Component of the cytoplasmic ribonucleoprotein (RNP) bodies which concentrate at the perinuclear region of the nurse cell and in punctate aggregates throughout the cytoplasm. Some cortical enrichment of these aggregates is also observed. At stage 7, accumulates in the anterior cytoplasm of the oocyte, whereas during stage 10 accumulates at the posterior pole. This posterior distribution is not maintained in later developmental stages. With evidence from 4 publications.
Nucleus Component of the cytoplasmic ribonucleoprotein (RNP) bodies which concentrate at the perinuclear region of the nurse cell and in punctate aggregates throughout the cytoplasm. Some cortical enrichment of these aggregates is also observed. At stage 7, accumulates in the anterior cytoplasm of the oocyte, whereas during stage 10 accumulates at the posterior pole. This posterior distribution is not maintained in later developmental stages. With evidence from 4 publications.
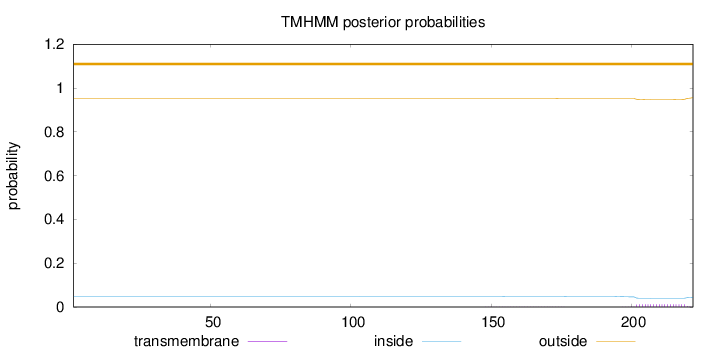
Length:
222
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24966
Exp number, first 60 AAs:
0.00039
Total prob of N-in:
0.04818
outside
1 - 222
Population Genetic Test Statistics
Pi
210.897855
Theta
166.138892
Tajima's D
0.77488
CLR
0
CSRT
0.592420378981051
Interpretation
Uncertain
