Gene
KWMTBOMO01354
Pre Gene Modal
BGIBMGA008895
Annotation
PREDICTED:_polypeptide_N-acetylgalactosaminyltransferase_3-like_[Bombyx_mori]
Full name
Polypeptide N-acetylgalactosaminyltransferase
+ More
Polypeptide N-acetylgalactosaminyltransferase 3
Polypeptide N-acetylgalactosaminyltransferase 3
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Protein-UDP acetylgalactosaminyltransferase 3
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 3
Protein-UDP acetylgalactosaminyltransferase 3
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 3
Location in the cell
PlasmaMembrane Reliability : 1.737
Sequence
CDS
ATGCTTCGGACCGAGTTTGGTGCTTTGTTATGTCGAGGACGTAAGAAAAGGATTGTGACATTGATACTAATATTGATATTTTTAATAAACGCTATGTTTTATGTTAGTTTTGAATTGTATAATACGGTTATGAGAAAAAATAAGAAGCTCTGGTATAACAGATTGAAGTATGATAAGAACTCATATGTTGATGAAAGTGGTATGAGAGTTATCGTCGGACATTATGTTGGTGGTATAGGAGGAGCGAATCTATCAGAAGAAATAATGCACACTAATAATTATTCACCAGTACCTGGAGCCGGAGAGGGTGGTAGACCTGTGCAGTTGTCTCCACGAGAGCTCATTACTGCTAGAGAGCTCTATACATTACATTCTTATAATATACTTGTTAGTGATAGGATAGCTATTAATAGAAGTCTTCCTGATATGCGAAGTGATAGTTGTAGAAGCGTCGTGTATGATACGGAAGAACTTCCGACTGCTAGTGTTATAATAGTCTTCCATAATGAGGCGTGGTCTACGTTAATGAGGACCATCATGTCAGTTCTGATGAGGTCCCCACAGCTTTTACTAAAGCAGATAATTCTTGTTGACGATGCGAGTGACAGGAGATATCTAAGTGAGGAATTGGATAACGCAGTAGCTAATTTAGACAAAGTGCAAATACTGAGGAGTTCAAACAGGAATGGTTTGGTTGGAGCCAGGTTGATGGGCGCCAGGGTTGCAACTGGAGATGTGTTAGTCTTTTTGGACGCACATTGTGAGGTTACAAAAGGATGGCTCGAGCCCCTATTGGATAGAGCAGGTAATGATGACGTTTTCATTTGTCCCCACATAGATCTATTATCAGAGGACACTTTAGCCTATACTAAAAGTATAGACGCCCACTGGGGTGCGTTCAGTTGGCGCTTACATTTCCGTTGGCTCACTCCGAGTGGAGATGTAATGTTAAAGAAGATTAGGAATCCGTCAAAACCGTATCCCACGCCGGCGATGGCGGGAGGATTGTTCGCAGTCAGAAAAAGTTTGTTCTGGAGATTGGGCGGTTATGATTCGGGTATGCTGATATGGGGCGCTGAGAACTTGGAGTTGTCGTGGAGGGCATGGCAGTGCGGTGCAAGGGTTGAAATAACGCCGTGCTCACGAGTCGGACATATATTCAGGAGACATAGTCCGTACAAATATCCAGGGGGTGTATCTAAAGTGTTGAACTCGAATTTGGCTCGCGCTGCCACAGTGTGGATGGACGAATGGGCAGATTTCTTCTTCAAATTCAACCCGCCAATTTCCGCGATACGCGATCAACAAAACGTTTACGATAGAGTGGAATTACGTAAGAATTTACACTGCAAAAGTTTCGAATGGTATTTAGAGAATGTGTGGCCGCAACACTTTTTTCCGACAACCAAGAGATGGTTTGGGCGAATCAAAAATGAAAAAGGAGCTTGTTTGGGTATTACTGGAGGAACACCAGGACTTGGAGCTCCGGCTTTGGGTCTGCAATGCGGTACTGACTTAGACCTTGATAGACTACTTGTCTATACTCCGGAAGGCAAGATAATGGCGGACGAGGGGCTGTGTTTGCAGCAAGGAAACGGCCGAGCCGTGTGGAAAAGTTGTAGTGACAATAAAAAGCAAATTTGGGTGCAAGATGGTGCTAGATTAAAAACTATAAGTGGTTCTTGTCTTACGTTGCTAATCTCCGAGAAAAATAGTGATGGTATTGATGAAGCACTAACGTCTAAGCGATGCGTTGAAGACGATAGTCAAATATGGCATTTTGAACGTGTGCCTTGGCGCTGA
Protein
MLRTEFGALLCRGRKKRIVTLILILIFLINAMFYVSFELYNTVMRKNKKLWYNRLKYDKNSYVDESGMRVIVGHYVGGIGGANLSEEIMHTNNYSPVPGAGEGGRPVQLSPRELITARELYTLHSYNILVSDRIAINRSLPDMRSDSCRSVVYDTEELPTASVIIVFHNEAWSTLMRTIMSVLMRSPQLLLKQIILVDDASDRRYLSEELDNAVANLDKVQILRSSNRNGLVGARLMGARVATGDVLVFLDAHCEVTKGWLEPLLDRAGNDDVFICPHIDLLSEDTLAYTKSIDAHWGAFSWRLHFRWLTPSGDVMLKKIRNPSKPYPTPAMAGGLFAVRKSLFWRLGGYDSGMLIWGAENLELSWRAWQCGARVEITPCSRVGHIFRRHSPYKYPGGVSKVLNSNLARAATVWMDEWADFFFKFNPPISAIRDQQNVYDRVELRKNLHCKSFEWYLENVWPQHFFPTTKRWFGRIKNEKGACLGITGGTPGLGAPALGLQCGTDLDLDRLLVYTPEGKIMADEGLCLQQGNGRAVWKSCSDNKKQIWVQDGARLKTISGSCLTLLISEKNSDGIDEALTSKRCVEDDSQIWHFERVPWR
Summary
Description
Catalyzes the initial reaction in O-linked oligosaccharide biosynthesis, the transfer of an N-acetyl-D-galactosamine residue to a serine or threonine residue on the protein receptor. It can both act as a peptide transferase that transfers GalNAc onto unmodified peptide substrates, and as a glycopeptide transferase that requires the prior addition of a GalNAc on a peptide before adding additional GalNAc moieties. Prefers EA2 as substrate. Has weak activity toward Muc5AC-3, -13 and -3/13 substrates. Plays a critical role in the regulation of integrin-mediated cell adhesion during wing development by influencing, via glycosylation, the secretion and localization of the integrin ligand Tig to the basal cell layer interface.
Catalytic Activity
L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-seryl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Glycosyltransferase
Golgi apparatus
Lectin
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Polypeptide N-acetylgalactosaminyltransferase
Uniprot
H9JH98
A0A212F596
A0A194PUF8
A0A2W1BV78
A0A2H1VGA6
A0A194RMI0
+ More
A0A1Y1MNF4 A0A067R7V4 D6WWQ3 A0A2J7RBU6 J9K3I4 A0A146LW32 A0A2S2P4H4 A0A0K8TBB6 A0A0C9QJW7 E0VX75 A0A0T6B172 A0A1W4XAW7 A0A026W442 A0A2H8TH72 A0A151J9S3 A0A158P1T7 A0A2S2R5Q9 A0A1B6CFV1 U4TZY3 A0A2A3EKJ3 A0A151XAL9 A0A0L7R5F8 A0A1B6EPV9 F4W6Q8 K7J884 A0A088APX4 A0A195CLS3 A0A195EQH4 A0A154P779 A0A195AWN0 A0A162Q9R2 A0A2P6KG00 A0A0P5YH87 A0A0P6FTD9 A0A0K2VIY1 A0A1S3DJY0 E2A7T4 T1HSC3 E2B2L3 A0A226EH74 E9H1K7 A0A0P4XQ53 A0A0P5KKA9 A0A0P5L8K0 A0A0M8ZQ52 A0A0P5MES9 A0A0A9YEM7 A0A183UCT4 T1L225 A0A0P5G6V0 A0A0N4VJP9 A0A0N8ASS8 A0A3B0JPI1 A0A1I7VTE4 A0A336KLY3 A0A182E8W0 T1JAK0 B4GCL2 Q292V0 A0A0P6HJM5 A0A0L0CCV9 B4P1D5 Q9Y117 A0A0M4EJ61
A0A1Y1MNF4 A0A067R7V4 D6WWQ3 A0A2J7RBU6 J9K3I4 A0A146LW32 A0A2S2P4H4 A0A0K8TBB6 A0A0C9QJW7 E0VX75 A0A0T6B172 A0A1W4XAW7 A0A026W442 A0A2H8TH72 A0A151J9S3 A0A158P1T7 A0A2S2R5Q9 A0A1B6CFV1 U4TZY3 A0A2A3EKJ3 A0A151XAL9 A0A0L7R5F8 A0A1B6EPV9 F4W6Q8 K7J884 A0A088APX4 A0A195CLS3 A0A195EQH4 A0A154P779 A0A195AWN0 A0A162Q9R2 A0A2P6KG00 A0A0P5YH87 A0A0P6FTD9 A0A0K2VIY1 A0A1S3DJY0 E2A7T4 T1HSC3 E2B2L3 A0A226EH74 E9H1K7 A0A0P4XQ53 A0A0P5KKA9 A0A0P5L8K0 A0A0M8ZQ52 A0A0P5MES9 A0A0A9YEM7 A0A183UCT4 T1L225 A0A0P5G6V0 A0A0N4VJP9 A0A0N8ASS8 A0A3B0JPI1 A0A1I7VTE4 A0A336KLY3 A0A182E8W0 T1JAK0 B4GCL2 Q292V0 A0A0P6HJM5 A0A0L0CCV9 B4P1D5 Q9Y117 A0A0M4EJ61
EC Number
2.4.1.-
2.4.1.41
2.4.1.41
Pubmed
EMBL
BABH01012709
AGBW02010217
OWR48907.1
KQ459591
KPI96957.1
KZ149894
+ More
PZC78879.1 ODYU01002421 SOQ39878.1 KQ459989 KPJ18732.1 GEZM01026154 JAV87204.1 KK852639 KDR19622.1 KQ971361 EFA08098.1 NEVH01005890 PNF38302.1 ABLF02017875 ABLF02017876 ABLF02017877 ABLF02017878 ABLF02017879 ABLF02045082 ABLF02060713 GDHC01013711 GDHC01013593 GDHC01008069 JAQ04918.1 JAQ05036.1 JAQ10560.1 GGMR01011701 MBY24320.1 GBRD01003120 JAG62701.1 GBYB01000902 JAG70669.1 DS235829 EEB17981.1 LJIG01016278 KRT81081.1 KK107436 QOIP01000002 EZA50822.1 RLU26044.1 GFXV01001247 MBW13052.1 KQ979361 KYN21820.1 ADTU01006849 GGMS01015867 MBY85070.1 GEDC01025083 GEDC01023436 GEDC01009517 GEDC01004538 JAS12215.1 JAS13862.1 JAS27781.1 JAS32760.1 KB631813 ERL86372.1 KZ288232 PBC31551.1 KQ982335 KYQ57423.1 KQ414654 KOC66059.1 GECZ01029810 JAS39959.1 GL887707 EGI70134.1 KQ977595 KYN01650.1 KQ982021 KYN30463.1 KQ434831 KZC07795.1 KQ976731 KYM76374.1 LRGB01000372 KZS19491.1 MWRG01013029 PRD25231.1 GDIP01057739 JAM45976.1 GDIQ01045819 JAN48918.1 HACA01032570 CDW49931.1 GL437384 EFN70514.1 ACPB03021813 GL445159 EFN90069.1 LNIX01000004 OXA56071.1 GL732583 EFX74488.1 GDIP01239683 JAI83718.1 GDIQ01184360 JAK67365.1 GDIQ01174606 JAK77119.1 KQ435915 KOX68825.1 GDIQ01156712 JAK95013.1 GBHO01034668 GBHO01034665 GBHO01013518 JAG08936.1 JAG08939.1 JAG30086.1 UYWY01019470 VDM37613.1 CAEY01000924 GDIQ01246436 JAK05289.1 UXUI01010807 VDD95644.1 GDIQ01246435 JAK05290.1 OUUW01000001 SPP73088.1 JH712080 EFO27036.2 UFQS01000613 UFQT01000613 SSX05397.1 SSX25758.1 UYRW01001012 VDK73133.1 JH432001 CH479181 EDW31460.1 CM000071 EAL24761.1 GDIQ01019131 JAN75606.1 JRES01000678 KNC29299.1 CM000157 EDW89137.1 AE013599 AF145655 CP012524 ALC41255.1
PZC78879.1 ODYU01002421 SOQ39878.1 KQ459989 KPJ18732.1 GEZM01026154 JAV87204.1 KK852639 KDR19622.1 KQ971361 EFA08098.1 NEVH01005890 PNF38302.1 ABLF02017875 ABLF02017876 ABLF02017877 ABLF02017878 ABLF02017879 ABLF02045082 ABLF02060713 GDHC01013711 GDHC01013593 GDHC01008069 JAQ04918.1 JAQ05036.1 JAQ10560.1 GGMR01011701 MBY24320.1 GBRD01003120 JAG62701.1 GBYB01000902 JAG70669.1 DS235829 EEB17981.1 LJIG01016278 KRT81081.1 KK107436 QOIP01000002 EZA50822.1 RLU26044.1 GFXV01001247 MBW13052.1 KQ979361 KYN21820.1 ADTU01006849 GGMS01015867 MBY85070.1 GEDC01025083 GEDC01023436 GEDC01009517 GEDC01004538 JAS12215.1 JAS13862.1 JAS27781.1 JAS32760.1 KB631813 ERL86372.1 KZ288232 PBC31551.1 KQ982335 KYQ57423.1 KQ414654 KOC66059.1 GECZ01029810 JAS39959.1 GL887707 EGI70134.1 KQ977595 KYN01650.1 KQ982021 KYN30463.1 KQ434831 KZC07795.1 KQ976731 KYM76374.1 LRGB01000372 KZS19491.1 MWRG01013029 PRD25231.1 GDIP01057739 JAM45976.1 GDIQ01045819 JAN48918.1 HACA01032570 CDW49931.1 GL437384 EFN70514.1 ACPB03021813 GL445159 EFN90069.1 LNIX01000004 OXA56071.1 GL732583 EFX74488.1 GDIP01239683 JAI83718.1 GDIQ01184360 JAK67365.1 GDIQ01174606 JAK77119.1 KQ435915 KOX68825.1 GDIQ01156712 JAK95013.1 GBHO01034668 GBHO01034665 GBHO01013518 JAG08936.1 JAG08939.1 JAG30086.1 UYWY01019470 VDM37613.1 CAEY01000924 GDIQ01246436 JAK05289.1 UXUI01010807 VDD95644.1 GDIQ01246435 JAK05290.1 OUUW01000001 SPP73088.1 JH712080 EFO27036.2 UFQS01000613 UFQT01000613 SSX05397.1 SSX25758.1 UYRW01001012 VDK73133.1 JH432001 CH479181 EDW31460.1 CM000071 EAL24761.1 GDIQ01019131 JAN75606.1 JRES01000678 KNC29299.1 CM000157 EDW89137.1 AE013599 AF145655 CP012524 ALC41255.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000027135
UP000007266
+ More
UP000235965 UP000007819 UP000009046 UP000192223 UP000053097 UP000279307 UP000078492 UP000005205 UP000030742 UP000242457 UP000075809 UP000053825 UP000007755 UP000002358 UP000005203 UP000078542 UP000078541 UP000076502 UP000078540 UP000076858 UP000079169 UP000000311 UP000015103 UP000008237 UP000198287 UP000000305 UP000053105 UP000050794 UP000267007 UP000015104 UP000038041 UP000274131 UP000268350 UP000095285 UP000077448 UP000271087 UP000008744 UP000001819 UP000037069 UP000002282 UP000000803 UP000092553
UP000235965 UP000007819 UP000009046 UP000192223 UP000053097 UP000279307 UP000078492 UP000005205 UP000030742 UP000242457 UP000075809 UP000053825 UP000007755 UP000002358 UP000005203 UP000078542 UP000078541 UP000076502 UP000078540 UP000076858 UP000079169 UP000000311 UP000015103 UP000008237 UP000198287 UP000000305 UP000053105 UP000050794 UP000267007 UP000015104 UP000038041 UP000274131 UP000268350 UP000095285 UP000077448 UP000271087 UP000008744 UP000001819 UP000037069 UP000002282 UP000000803 UP000092553
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JH98
A0A212F596
A0A194PUF8
A0A2W1BV78
A0A2H1VGA6
A0A194RMI0
+ More
A0A1Y1MNF4 A0A067R7V4 D6WWQ3 A0A2J7RBU6 J9K3I4 A0A146LW32 A0A2S2P4H4 A0A0K8TBB6 A0A0C9QJW7 E0VX75 A0A0T6B172 A0A1W4XAW7 A0A026W442 A0A2H8TH72 A0A151J9S3 A0A158P1T7 A0A2S2R5Q9 A0A1B6CFV1 U4TZY3 A0A2A3EKJ3 A0A151XAL9 A0A0L7R5F8 A0A1B6EPV9 F4W6Q8 K7J884 A0A088APX4 A0A195CLS3 A0A195EQH4 A0A154P779 A0A195AWN0 A0A162Q9R2 A0A2P6KG00 A0A0P5YH87 A0A0P6FTD9 A0A0K2VIY1 A0A1S3DJY0 E2A7T4 T1HSC3 E2B2L3 A0A226EH74 E9H1K7 A0A0P4XQ53 A0A0P5KKA9 A0A0P5L8K0 A0A0M8ZQ52 A0A0P5MES9 A0A0A9YEM7 A0A183UCT4 T1L225 A0A0P5G6V0 A0A0N4VJP9 A0A0N8ASS8 A0A3B0JPI1 A0A1I7VTE4 A0A336KLY3 A0A182E8W0 T1JAK0 B4GCL2 Q292V0 A0A0P6HJM5 A0A0L0CCV9 B4P1D5 Q9Y117 A0A0M4EJ61
A0A1Y1MNF4 A0A067R7V4 D6WWQ3 A0A2J7RBU6 J9K3I4 A0A146LW32 A0A2S2P4H4 A0A0K8TBB6 A0A0C9QJW7 E0VX75 A0A0T6B172 A0A1W4XAW7 A0A026W442 A0A2H8TH72 A0A151J9S3 A0A158P1T7 A0A2S2R5Q9 A0A1B6CFV1 U4TZY3 A0A2A3EKJ3 A0A151XAL9 A0A0L7R5F8 A0A1B6EPV9 F4W6Q8 K7J884 A0A088APX4 A0A195CLS3 A0A195EQH4 A0A154P779 A0A195AWN0 A0A162Q9R2 A0A2P6KG00 A0A0P5YH87 A0A0P6FTD9 A0A0K2VIY1 A0A1S3DJY0 E2A7T4 T1HSC3 E2B2L3 A0A226EH74 E9H1K7 A0A0P4XQ53 A0A0P5KKA9 A0A0P5L8K0 A0A0M8ZQ52 A0A0P5MES9 A0A0A9YEM7 A0A183UCT4 T1L225 A0A0P5G6V0 A0A0N4VJP9 A0A0N8ASS8 A0A3B0JPI1 A0A1I7VTE4 A0A336KLY3 A0A182E8W0 T1JAK0 B4GCL2 Q292V0 A0A0P6HJM5 A0A0L0CCV9 B4P1D5 Q9Y117 A0A0M4EJ61
PDB
1XHB
E-value=6.94638e-99,
Score=923
Ontologies
PATHWAY
GO
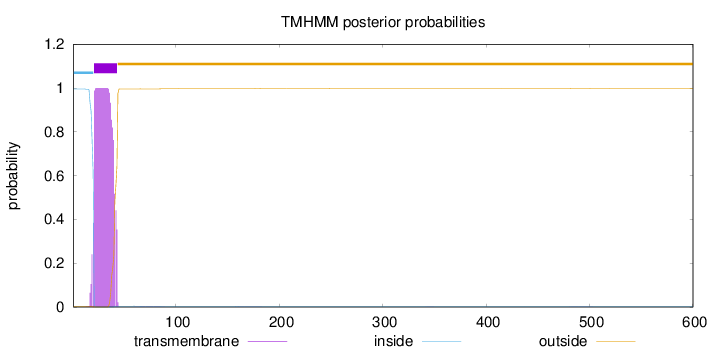
Topology
Subcellular location
Golgi apparatus membrane
Length:
600
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.5396699999999
Exp number, first 60 AAs:
21.46449
Total prob of N-in:
0.99607
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 600
Population Genetic Test Statistics
Pi
230.679969
Theta
174.29066
Tajima's D
0.847638
CLR
0.709687
CSRT
0.614619269036548
Interpretation
Uncertain
