Pre Gene Modal
BGIBMGA008997
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_DHX36_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.134
Sequence
CDS
ATGTATTATAAAGCTTTAAGTATTGAAAAAGGAAAGAAAAATAAGAAAAACAATGTGATACGCTTCAAAATTCCACCAAGCATTGTCTCATCACTGTATAGTAATTTAGCATCAATAGAAAAGCTTGCTAAAGGACTGAACATTTGCATAAAACCAATAGAAAAATTGACTATTGCAGAAACACCAAAAGGCTTAAAACCTGAACTGTCATTACATGAAGCGATGTACATGAACACGAGTACAGCTGAAAACAAAAACAGTGTGCAGGCAAGTGATAATGAAAATCTATTTGAAGCAAAAACACCACTAACTGATTTTAAAAAGCTAGAAGATATTTCAATGATATCTGAAGATGCTGAATTACGTTCACCAAGCGATGTTGACAACAAATCAAATGGGCCATCTAAGGATTTTATTTCAGTAAAGGATGAAACTGTGGAAATCAAATTAGAGAACCGTTTTTTATCACTTAGAAGTTCAGTTAACTATAAATATGGCTACGAAGATATCATAACTGGGACGTTTGATGAAAAATTAGATGAATGCTTGGCAAGAGGAATCAACATTAATATTAAGAATGAAGTAAATAATGATCTCAGTTTTGCAATGATAGAAGAATACAATGATATGTTGAGCAAAGATGGTTATAAATTGCACATGAAATTTCGTGACAAGCTACCTACATACCAAAAATATGAAGAGCTCTTAAACATGATCAACAATAATCAAGTTTTGGTCATAAGTGGAGAAACAGGATGTGGCAAATCGACACAGGTACCTCAAATAATTCTTGATGATGCCATTATTAACAAAAGAGGTGCAAATGTGAAGATTCTAGTAACTCAACCCAGGAGAATCGCAGCTTCATCATTAGCTATGAGAGTGGCTGAAGAGAGAAATGAGAAACTAGGGAATAGTGTGGGATATGCTGTTAGATTGGAGAAGGTTGATCCAAGGCCTCTAGGAAGCATTATGTTTTGTACCACAGGTATACTACTGACTGAACTTGAAGTTAATCAAAGTTTGTCTAACTATAGCCATATTATCCTCGATGAAGTGCATGAAAGGGACACACATATCGACTTGTCTATGTGTATGCTAAAAAAGGTACTGAATAAGCGAAAAGACTTGAAACTAATACTTATGAGTGCCACAATAAATGCTGAACATCTCACTTCCTACTTTGGTGACTGTCAAAGAATCCACATTGAAGGTATAGCATATCCTGTCCAAGATGTTTATTTAGAAGAGATATTGAAAATGACTGGATATAAGTTGCCACCCGCTGAAAACAAAAGAGCTGATAAGAGAAAGACTTGGCAAAATCACACGAAAAAGGCTAAAGCTATGGCAGCCTCTGAAATGGAAAAAGACATACAATATAAAGCAGAAATTGGACCATGGCTAGAAACCGTAAAAGGCAAAATCGGTTGGGATGTATGTACTACATTACTAGATGCAAGGATCGAAGATATTAATATTGATTTAATTGTAGAAGTTCTTAAATTAATTAGCAGTAAAGAACCCGGCGCCGTCCTGGTGTTTCTACCCGGAATCGGTGATATAACCAAGCTCATGTCCGTGATGGAGCAGAGTTGTGAATTTCCGAAATCGAAATTCGACGTATATCCTCTACATTCGCGACTACCTTCTTTGGAACAACACAAGATTTTCCAAAGACCACCGACCGGTATCCGAAAAATCATAATAGCTACGAATATCGCGGAAACATCAATTACAATCGATGATATTGTATACGTAATCGATTGCGGTAAGATTAAATACAGCGGACTAAGCATTGCGGACAACATATCGACATTGAAAACTGAGTGGATATCTCAGGCCAATTTGCGACAAAGACGAGGCCGCGCCGGGCGATGCCAACCCGGCGTCTGTTATCACTTAGTTACATCGTTCCGAGCTGGGAAACTCGAACCGAGCCTAAAGCCTGAGATGCAGAGGAGTGACCTACTCGAGCCCGTGCTTGCCATAAAACGGCTTCGACTCGGAAAAGCTGCTGATGCTTTAAAGGATATGGCGGCTCCACCGGCAGAAGAAACTATTAAAGCTGCCGTTAAACATTTACAACAATGTGGAGCCCTCAATGAATCAGAAATCTTAACACCTCTTGGTTGGCATTTGGCACGACTACCAGTTCACCCTGCGGCTGGTAAATTATTGCTGTTGGGAGCGCTGTTCGGTTGTTTGGACCGAGCCGCCAGCGTTGCCGCCGTTTGGGGCTTCAAAGACCCTTTTCAGTTAGTTATTGGAAAAGAAAAACAAATGGACCTAGCAAAAAAGGAATTAGCCCTAGGTGAGCCAAGTGACCACGTTGCGATATCCGAGGCAATTATACAATGGGAACAATGCCCCAGGCAACATCGTAATAATTTCGCATACAAAAACTTCCTTTCCATGAATACTCTGAAATTACTCTCTGAAATGAAGCATCAGCTGGCCGACAATTTGAGGCAGATGGGCTTTTTGAATACTGGCAACATCAAATCGGCTTGGAAAAATAGAAATGCTAATAATCTAAGCCTTTTTAAAGCTATAGTCGCTGCAGCGCTATACCCCAATATTGGTTCTGTGAGGTGGACGAATCTTCATAACCGAAATCCACGTAAGACTCCCCGCATCAAGGTTTGGACTCCGGAAGACGGCTCGATCGCGCTGCACCCCAGCAGCGTCATGGCGAACAAACACAACGGCGGCCTCAACGCTCAGCCGATGTGTGCGAGCCCGGGCGCTAACTGGCTGGTGTACTGGTTGAAGCGGCGCTCGACCGATTTGTTCTTGTTTGACGTCACTCTGGTCTATACTCTGCCTCTGTTGTTTTTCGGTGAACTTCACGTGTCTCCAGCAGCTGAAGACCCAGATGACTGTATCATATCGATAGAAAAGATATCTGTTTGTTGCAAGAAGGAAACTACACAATTGCTGTTTGAAATGCGGTCGTTATTAGACCACGTTTTAGCCGACAAGATCATGGCGTCTTCCGAACACTCCGTAAAAAATAATGCCTTTGAAGAACAGGTTCTGAATGCAGTGATAAAGCTTATTACAGCAGAGGACGAGGGCGCTGAATATTTGAACAGCGACGACGACAATTCCGAATCTGATCATTCAGAATACGAAACGAGCAGGAGGTGGTACCGTTAA
Protein
MYYKALSIEKGKKNKKNNVIRFKIPPSIVSSLYSNLASIEKLAKGLNICIKPIEKLTIAETPKGLKPELSLHEAMYMNTSTAENKNSVQASDNENLFEAKTPLTDFKKLEDISMISEDAELRSPSDVDNKSNGPSKDFISVKDETVEIKLENRFLSLRSSVNYKYGYEDIITGTFDEKLDECLARGININIKNEVNNDLSFAMIEEYNDMLSKDGYKLHMKFRDKLPTYQKYEELLNMINNNQVLVISGETGCGKSTQVPQIILDDAIINKRGANVKILVTQPRRIAASSLAMRVAEERNEKLGNSVGYAVRLEKVDPRPLGSIMFCTTGILLTELEVNQSLSNYSHIILDEVHERDTHIDLSMCMLKKVLNKRKDLKLILMSATINAEHLTSYFGDCQRIHIEGIAYPVQDVYLEEILKMTGYKLPPAENKRADKRKTWQNHTKKAKAMAASEMEKDIQYKAEIGPWLETVKGKIGWDVCTTLLDARIEDINIDLIVEVLKLISSKEPGAVLVFLPGIGDITKLMSVMEQSCEFPKSKFDVYPLHSRLPSLEQHKIFQRPPTGIRKIIIATNIAETSITIDDIVYVIDCGKIKYSGLSIADNISTLKTEWISQANLRQRRGRAGRCQPGVCYHLVTSFRAGKLEPSLKPEMQRSDLLEPVLAIKRLRLGKAADALKDMAAPPAEETIKAAVKHLQQCGALNESEILTPLGWHLARLPVHPAAGKLLLLGALFGCLDRAASVAAVWGFKDPFQLVIGKEKQMDLAKKELALGEPSDHVAISEAIIQWEQCPRQHRNNFAYKNFLSMNTLKLLSEMKHQLADNLRQMGFLNTGNIKSAWKNRNANNLSLFKAIVAAALYPNIGSVRWTNLHNRNPRKTPRIKVWTPEDGSIALHPSSVMANKHNGGLNAQPMCASPGANWLVYWLKRRSTDLFLFDVTLVYTLPLLFFGELHVSPAAEDPDDCIISIEKISVCCKKETTQLLFEMRSLLDHVLADKIMASSEHSVKNNAFEEQVLNAVIKLITAEDEGAEYLNSDDDNSESDHSEYETSRRWYR
Summary
Uniprot
H9JHJ9
A0A2H1VDY7
A0A212F590
A0A194PVD9
A0A2W1C175
S4PXP2
+ More
A0A0L7KTG9 E2AXE3 A0A195FSJ8 A0A2A3ECJ0 A0A088A6K8 A0A154PD24 F4WBQ2 A0A232ETT4 K7IVU8 A0A2J7RBV0 A0A2J7RBV4 A0A0L7R8J1 A0A151XEP8 A0A195BD28 A0A195DI87 E9IG17 A0A0N0U387 A0A067R851 A0A1W4XL65 A0A2J7RBW7 A0A158NRK7 A0A069DXN1 A0A224X6C8 A0A023F1I7 A0A3M6TB07 A0A087XVI7 A0A2B4RQD7 M3ZH26 A0A3B3VIA1 A0A3Q0HJ72 A0A1L8GAK8 A0A3P9K5Z8 A0A3P9N3T6 A0A3B3HH74 A0A3B3YE48 A0A1A8QCW6 A0A1A8NGH4 A0A1A8KW15 A0A3P9HF04 A0A131YN63 A0A1A8CX65 A0A1A8IEX4 A0A1A8QZ52 A0A1A8ML94 A0A1A8B508 A0A1A8TYY7 A0A1A7WZJ2 A0A1A7YRG5 A0A131XYB7 A0A1A8FJ48 A0A147BQ62 A0A093KUT0 A0A091IEM5 A0A2I0M1J8 H3B943 A0A0T6BCB5 H3B942 A0A3B3SCT4 A0A0A0AF48 D2HQH3 F6Q4S1 A0A1X7VIU1 A0A091W2D4 A0A093GU86 M3W2Z7 A0A093SNM1 A0A1V4KM25 A0A093NBF9 F7ETR2 A0A091SHM9 G1LZ43 A0A1Y1N3Q9 A0A146ZY80 A0A091MV85 A0A091GIZ3 A0A3P8XVN2 A0A091IXG9 A0A091P0W0 U3IYV0 A0A384DDN6 A0A2K5C9F2 A0A091THF0 A0A3P9AWJ8 A0A2K6T8G7 A0A3B4VPF7 A0A1S2ZLC6 A0A2U3W9Q0 F6X412 A0A091HEQ4 W5M293 A0A091XVC9 I3J6H4 W5M2A6 A0A087QIK8
A0A0L7KTG9 E2AXE3 A0A195FSJ8 A0A2A3ECJ0 A0A088A6K8 A0A154PD24 F4WBQ2 A0A232ETT4 K7IVU8 A0A2J7RBV0 A0A2J7RBV4 A0A0L7R8J1 A0A151XEP8 A0A195BD28 A0A195DI87 E9IG17 A0A0N0U387 A0A067R851 A0A1W4XL65 A0A2J7RBW7 A0A158NRK7 A0A069DXN1 A0A224X6C8 A0A023F1I7 A0A3M6TB07 A0A087XVI7 A0A2B4RQD7 M3ZH26 A0A3B3VIA1 A0A3Q0HJ72 A0A1L8GAK8 A0A3P9K5Z8 A0A3P9N3T6 A0A3B3HH74 A0A3B3YE48 A0A1A8QCW6 A0A1A8NGH4 A0A1A8KW15 A0A3P9HF04 A0A131YN63 A0A1A8CX65 A0A1A8IEX4 A0A1A8QZ52 A0A1A8ML94 A0A1A8B508 A0A1A8TYY7 A0A1A7WZJ2 A0A1A7YRG5 A0A131XYB7 A0A1A8FJ48 A0A147BQ62 A0A093KUT0 A0A091IEM5 A0A2I0M1J8 H3B943 A0A0T6BCB5 H3B942 A0A3B3SCT4 A0A0A0AF48 D2HQH3 F6Q4S1 A0A1X7VIU1 A0A091W2D4 A0A093GU86 M3W2Z7 A0A093SNM1 A0A1V4KM25 A0A093NBF9 F7ETR2 A0A091SHM9 G1LZ43 A0A1Y1N3Q9 A0A146ZY80 A0A091MV85 A0A091GIZ3 A0A3P8XVN2 A0A091IXG9 A0A091P0W0 U3IYV0 A0A384DDN6 A0A2K5C9F2 A0A091THF0 A0A3P9AWJ8 A0A2K6T8G7 A0A3B4VPF7 A0A1S2ZLC6 A0A2U3W9Q0 F6X412 A0A091HEQ4 W5M293 A0A091XVC9 I3J6H4 W5M2A6 A0A087QIK8
Pubmed
19121390
22118469
26354079
28756777
23622113
26227816
+ More
20798317 21719571 28648823 20075255 21282665 24845553 21347285 26334808 25474469 30382153 23542700 27762356 17554307 26830274 29652888 23371554 9215903 29240929 20010809 19892987 17975172 20431018 28004739 25069045 23749191 24813606 25186727
20798317 21719571 28648823 20075255 21282665 24845553 21347285 26334808 25474469 30382153 23542700 27762356 17554307 26830274 29652888 23371554 9215903 29240929 20010809 19892987 17975172 20431018 28004739 25069045 23749191 24813606 25186727
EMBL
BABH01012710
BABH01012711
ODYU01002049
SOQ39080.1
AGBW02010217
OWR48906.1
+ More
KQ459591 KPI96958.1 KZ149894 PZC78880.1 GAIX01004693 JAA87867.1 JTDY01005839 KOB66562.1 GL443548 EFN61923.1 KQ981280 KYN43416.1 KZ288300 PBC28896.1 KQ434874 KZC09687.1 GL888066 EGI68330.1 NNAY01002229 OXU21761.1 NEVH01005890 PNF38309.1 PNF38312.1 KQ414632 KOC67148.1 KQ982244 KYQ58849.1 KQ976511 KYM82476.1 KQ980824 KYN12603.1 GL762910 EFZ20446.1 KQ435922 KOX68608.1 KK852639 KDR19628.1 PNF38308.1 ADTU01023989 GBGD01000308 JAC88581.1 GFTR01008420 JAW08006.1 GBBI01003554 JAC15158.1 RCHS01003989 RMX38533.1 AYCK01011525 LSMT01000394 PFX18760.1 CM004474 OCT80873.1 HAEI01005008 SBR91366.1 HAEG01002811 SBR68001.1 HAEE01016675 SBR36725.1 GEDV01008575 JAP79982.1 HADZ01019320 SBP83261.1 HAED01009421 SBQ95633.1 HAEH01013780 SBR98294.1 HAEF01016182 SBR57341.1 HADY01024097 SBP62582.1 HAEJ01000059 SBS40516.1 HADW01009937 SBP11337.1 HADX01010559 SBP32791.1 GEFM01004534 JAP71262.1 HAEB01011632 SBQ58159.1 GEGO01002507 JAR92897.1 KK592723 KFW01181.1 KL218477 KFP05800.1 AKCR02000049 PKK23555.1 AFYH01073792 AFYH01073793 AFYH01073794 AFYH01073795 LJIG01001964 KRT84976.1 KL871660 KGL92781.1 GL193180 EFB20450.1 KL410321 KFQ95988.1 KK375638 KFV45816.1 AANG04001816 KL672030 KFW84209.1 LSYS01002834 OPJ85451.1 KL224626 KFW62288.1 AAMC01020934 AAMC01020935 KK473021 KFQ58124.1 ACTA01068082 ACTA01076082 GEZM01017017 JAV90966.1 GCES01015019 JAR71304.1 KK517980 KFP65405.1 KL448192 KFO81164.1 KK501044 KFP12278.1 KK647933 KFQ00053.1 ADON01099790 KK453376 KFQ74908.1 KL532613 KFO93302.1 AHAT01022488 KK736027 KFR16841.1 AERX01035202 KL225635 KFM01062.1
KQ459591 KPI96958.1 KZ149894 PZC78880.1 GAIX01004693 JAA87867.1 JTDY01005839 KOB66562.1 GL443548 EFN61923.1 KQ981280 KYN43416.1 KZ288300 PBC28896.1 KQ434874 KZC09687.1 GL888066 EGI68330.1 NNAY01002229 OXU21761.1 NEVH01005890 PNF38309.1 PNF38312.1 KQ414632 KOC67148.1 KQ982244 KYQ58849.1 KQ976511 KYM82476.1 KQ980824 KYN12603.1 GL762910 EFZ20446.1 KQ435922 KOX68608.1 KK852639 KDR19628.1 PNF38308.1 ADTU01023989 GBGD01000308 JAC88581.1 GFTR01008420 JAW08006.1 GBBI01003554 JAC15158.1 RCHS01003989 RMX38533.1 AYCK01011525 LSMT01000394 PFX18760.1 CM004474 OCT80873.1 HAEI01005008 SBR91366.1 HAEG01002811 SBR68001.1 HAEE01016675 SBR36725.1 GEDV01008575 JAP79982.1 HADZ01019320 SBP83261.1 HAED01009421 SBQ95633.1 HAEH01013780 SBR98294.1 HAEF01016182 SBR57341.1 HADY01024097 SBP62582.1 HAEJ01000059 SBS40516.1 HADW01009937 SBP11337.1 HADX01010559 SBP32791.1 GEFM01004534 JAP71262.1 HAEB01011632 SBQ58159.1 GEGO01002507 JAR92897.1 KK592723 KFW01181.1 KL218477 KFP05800.1 AKCR02000049 PKK23555.1 AFYH01073792 AFYH01073793 AFYH01073794 AFYH01073795 LJIG01001964 KRT84976.1 KL871660 KGL92781.1 GL193180 EFB20450.1 KL410321 KFQ95988.1 KK375638 KFV45816.1 AANG04001816 KL672030 KFW84209.1 LSYS01002834 OPJ85451.1 KL224626 KFW62288.1 AAMC01020934 AAMC01020935 KK473021 KFQ58124.1 ACTA01068082 ACTA01076082 GEZM01017017 JAV90966.1 GCES01015019 JAR71304.1 KK517980 KFP65405.1 KL448192 KFO81164.1 KK501044 KFP12278.1 KK647933 KFQ00053.1 ADON01099790 KK453376 KFQ74908.1 KL532613 KFO93302.1 AHAT01022488 KK736027 KFR16841.1 AERX01035202 KL225635 KFM01062.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000000311
UP000078541
+ More
UP000242457 UP000005203 UP000076502 UP000007755 UP000215335 UP000002358 UP000235965 UP000053825 UP000075809 UP000078540 UP000078492 UP000053105 UP000027135 UP000192223 UP000005205 UP000275408 UP000028760 UP000225706 UP000002852 UP000261500 UP000189705 UP000186698 UP000265180 UP000242638 UP000001038 UP000261480 UP000265200 UP000054308 UP000053872 UP000008672 UP000261540 UP000053858 UP000002281 UP000007879 UP000053283 UP000011712 UP000053258 UP000190648 UP000054081 UP000008143 UP000008912 UP000053760 UP000265140 UP000053119 UP000016666 UP000261680 UP000291021 UP000233020 UP000265160 UP000233220 UP000261420 UP000079721 UP000245340 UP000008225 UP000018468 UP000053605 UP000005207 UP000053286
UP000242457 UP000005203 UP000076502 UP000007755 UP000215335 UP000002358 UP000235965 UP000053825 UP000075809 UP000078540 UP000078492 UP000053105 UP000027135 UP000192223 UP000005205 UP000275408 UP000028760 UP000225706 UP000002852 UP000261500 UP000189705 UP000186698 UP000265180 UP000242638 UP000001038 UP000261480 UP000265200 UP000054308 UP000053872 UP000008672 UP000261540 UP000053858 UP000002281 UP000007879 UP000053283 UP000011712 UP000053258 UP000190648 UP000054081 UP000008143 UP000008912 UP000053760 UP000265140 UP000053119 UP000016666 UP000261680 UP000291021 UP000233020 UP000265160 UP000233220 UP000261420 UP000079721 UP000245340 UP000008225 UP000018468 UP000053605 UP000005207 UP000053286
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JHJ9
A0A2H1VDY7
A0A212F590
A0A194PVD9
A0A2W1C175
S4PXP2
+ More
A0A0L7KTG9 E2AXE3 A0A195FSJ8 A0A2A3ECJ0 A0A088A6K8 A0A154PD24 F4WBQ2 A0A232ETT4 K7IVU8 A0A2J7RBV0 A0A2J7RBV4 A0A0L7R8J1 A0A151XEP8 A0A195BD28 A0A195DI87 E9IG17 A0A0N0U387 A0A067R851 A0A1W4XL65 A0A2J7RBW7 A0A158NRK7 A0A069DXN1 A0A224X6C8 A0A023F1I7 A0A3M6TB07 A0A087XVI7 A0A2B4RQD7 M3ZH26 A0A3B3VIA1 A0A3Q0HJ72 A0A1L8GAK8 A0A3P9K5Z8 A0A3P9N3T6 A0A3B3HH74 A0A3B3YE48 A0A1A8QCW6 A0A1A8NGH4 A0A1A8KW15 A0A3P9HF04 A0A131YN63 A0A1A8CX65 A0A1A8IEX4 A0A1A8QZ52 A0A1A8ML94 A0A1A8B508 A0A1A8TYY7 A0A1A7WZJ2 A0A1A7YRG5 A0A131XYB7 A0A1A8FJ48 A0A147BQ62 A0A093KUT0 A0A091IEM5 A0A2I0M1J8 H3B943 A0A0T6BCB5 H3B942 A0A3B3SCT4 A0A0A0AF48 D2HQH3 F6Q4S1 A0A1X7VIU1 A0A091W2D4 A0A093GU86 M3W2Z7 A0A093SNM1 A0A1V4KM25 A0A093NBF9 F7ETR2 A0A091SHM9 G1LZ43 A0A1Y1N3Q9 A0A146ZY80 A0A091MV85 A0A091GIZ3 A0A3P8XVN2 A0A091IXG9 A0A091P0W0 U3IYV0 A0A384DDN6 A0A2K5C9F2 A0A091THF0 A0A3P9AWJ8 A0A2K6T8G7 A0A3B4VPF7 A0A1S2ZLC6 A0A2U3W9Q0 F6X412 A0A091HEQ4 W5M293 A0A091XVC9 I3J6H4 W5M2A6 A0A087QIK8
A0A0L7KTG9 E2AXE3 A0A195FSJ8 A0A2A3ECJ0 A0A088A6K8 A0A154PD24 F4WBQ2 A0A232ETT4 K7IVU8 A0A2J7RBV0 A0A2J7RBV4 A0A0L7R8J1 A0A151XEP8 A0A195BD28 A0A195DI87 E9IG17 A0A0N0U387 A0A067R851 A0A1W4XL65 A0A2J7RBW7 A0A158NRK7 A0A069DXN1 A0A224X6C8 A0A023F1I7 A0A3M6TB07 A0A087XVI7 A0A2B4RQD7 M3ZH26 A0A3B3VIA1 A0A3Q0HJ72 A0A1L8GAK8 A0A3P9K5Z8 A0A3P9N3T6 A0A3B3HH74 A0A3B3YE48 A0A1A8QCW6 A0A1A8NGH4 A0A1A8KW15 A0A3P9HF04 A0A131YN63 A0A1A8CX65 A0A1A8IEX4 A0A1A8QZ52 A0A1A8ML94 A0A1A8B508 A0A1A8TYY7 A0A1A7WZJ2 A0A1A7YRG5 A0A131XYB7 A0A1A8FJ48 A0A147BQ62 A0A093KUT0 A0A091IEM5 A0A2I0M1J8 H3B943 A0A0T6BCB5 H3B942 A0A3B3SCT4 A0A0A0AF48 D2HQH3 F6Q4S1 A0A1X7VIU1 A0A091W2D4 A0A093GU86 M3W2Z7 A0A093SNM1 A0A1V4KM25 A0A093NBF9 F7ETR2 A0A091SHM9 G1LZ43 A0A1Y1N3Q9 A0A146ZY80 A0A091MV85 A0A091GIZ3 A0A3P8XVN2 A0A091IXG9 A0A091P0W0 U3IYV0 A0A384DDN6 A0A2K5C9F2 A0A091THF0 A0A3P9AWJ8 A0A2K6T8G7 A0A3B4VPF7 A0A1S2ZLC6 A0A2U3W9Q0 F6X412 A0A091HEQ4 W5M293 A0A091XVC9 I3J6H4 W5M2A6 A0A087QIK8
PDB
5VHE
E-value=5.71519e-164,
Score=1486
Ontologies
GO
GO:0005524
GO:0004386
GO:0003676
GO:0016021
GO:0016787
GO:0005829
GO:0005634
GO:0045995
GO:0048027
GO:0000781
GO:0003725
GO:0034459
GO:0044806
GO:0051891
GO:0004003
GO:0000978
GO:0000287
GO:0060261
GO:0009615
GO:0016607
GO:0034644
GO:0090669
GO:0042826
GO:0003723
GO:1901534
GO:0002151
GO:0034605
GO:0061158
GO:1902741
GO:0003697
GO:1904358
GO:0002735
GO:0043123
GO:0035925
GO:0010494
GO:0010501
GO:0070034
GO:2000767
GO:1900153
GO:1902064
GO:0001503
GO:0043330
GO:0051880
GO:0006359
GO:0031442
GO:1903843
GO:0004004
GO:0008026
GO:0016820
GO:0016042
GO:0006334
GO:0003779
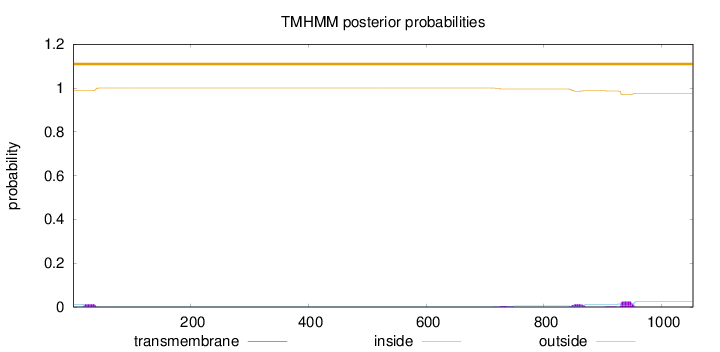
Topology
Length:
1053
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.12839
Exp number, first 60 AAs:
0.23842
Total prob of N-in:
0.01221
outside
1 - 1053
Population Genetic Test Statistics
Pi
257.138084
Theta
187.594058
Tajima's D
0.431369
CLR
1.114548
CSRT
0.498425078746063
Interpretation
Uncertain
