Gene
KWMTBOMO01351 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008894
Annotation
PREDICTED:_ADP-ribosylation_factor_2_[Plutella_xylostella]
Full name
ADP-ribosylation factor 2
Alternative Name
dARF II
Location in the cell
Mitochondrial Reliability : 1.053
Sequence
CDS
ATGGGTCTAACAATATCAAGTGTATTTACTAGATTGTTCGGCAAAAAACAAATGCGTATTCTTATGGTTGGTTTAGATGCTGCCGGAAAAACTACCATACTTTACAAATTAAAACTCGGTGAAATCGTGACTACGATACCGACAATCGGTTTCAATGTGGAAACAGTTGAGTACAAAAATATTAGTTTCACCGTGTGGGACGTTGGCGGCCAAGACAAAATCAGGCCTCTTTGGCGTCATTATTACCAGAACACTCAAGGCTTGATATTTGTTGTAGATTCTAGTGACAGAAAAAGAATAGCAGAAGCCGAAAATGAGTTGGCTAATATGTTAAAAGAAGATGAGCTTAGAGATGCAGTTATTTTAGTTTTTGCTAATAAACAAGACATGCCAAACGCGATGACTGCTGCAGAACTTACGAATGCCCTAAACCTTGGTAATCTGAGAAATCGACGGTGGTACATCCAAGCAACATGCGCTACACAAGGACAGGGGCTTTATGAAGGTCTTGATTGGCTGTCCAATGAGCTAGCGAAGAAATGA
Protein
MGLTISSVFTRLFGKKQMRILMVGLDAAGKTTILYKLKLGEIVTTIPTIGFNVETVEYKNISFTVWDVGGQDKIRPLWRHYYQNTQGLIFVVDSSDRKRIAEAENELANMLKEDELRDAVILVFANKQDMPNAMTAAELTNALNLGNLRNRRWYIQATCATQGQGLYEGLDWLSNELAKK
Summary
Description
GTP-binding protein involved in protein trafficking; may modulate vesicle budding and uncoating within the Golgi apparatus.
Similarity
Belongs to the small GTPase superfamily. Arf family.
Keywords
Complete proteome
ER-Golgi transport
Golgi apparatus
GTP-binding
Lipoprotein
Myristate
Nucleotide-binding
Protein transport
Reference proteome
Transport
Feature
chain ADP-ribosylation factor 2
Uniprot
S4PLM4
A0A1E1WR24
A0A2W1BUX3
A0A2A4ISZ1
A0A2H1VE15
A0A194RN27
+ More
I4DNX6 A0A212F5A4 A0A1D1V306 A0A336L2J2 T1DQ43 A0A182XV13 A0A2M3Z1B8 A0A2M4A499 A0A182M7Z2 A0A182QYJ0 A0A182T456 A0A182NLH7 W5JMB5 A0A182VRR2 A0A084W573 A0A182FNS8 A0A182RPB6 T1D515 A0A023EID5 A0A1Q3FBT4 B0W8N3 A0A182JHE6 E1ZVI0 A0A154PPJ6 A0A182JXQ5 F4WYZ2 A0A195BXZ2 A0A195FAS5 A0A158NC85 A0A151WFJ5 A0A0J7L480 A0A0P4WHL6 U5EXH1 A0A0M8ZQA2 E9ILY4 A0A1B6KPZ2 A0A2A3ED51 V9II32 A0A088A7D1 E2BQ20 A0A1B6F1U5 A0A182V8G9 A0A182TJK9 A0A182XHF6 A0A182LG43 Q7PQY7 A0A182HTZ0 A0A0L7R2Z1 A0A195EAB7 A0A026WVX0 D3IWC8 A0A182NZD7 A0A1W4XFG9 A0A097PIE9 A0A067R700 A0A1W4W163 A0A1L8DW98 L0MNA8 B4IKR6 B4R185 P40945 A0A0K8TNS0 A8C9W7 B3MZ91 B4NHH1 B3P9P3 B4L7F7 B4MF16 A0A0N8D229 A0A2H8TP42 Q1W9N5 A0A1Y1N522 Q29CQ9 B4H9Q6 A0A0M4ES39 A0A0P5W4U8 B4PW48 A0A2P2HYT8 A0A0L0C9F7 W8BX92 A0A0K8UWR1 A0A310SM75 A0A2R5LEQ3 A0A293N4R9 E9HDF7 A0A1D2N474 A0A0P6I372 A0A1W7RB47 H9JH97 R7U3Y2 B4JZR6 A0A139WNV7 Q6P5M5 A0A2I9LNP6
I4DNX6 A0A212F5A4 A0A1D1V306 A0A336L2J2 T1DQ43 A0A182XV13 A0A2M3Z1B8 A0A2M4A499 A0A182M7Z2 A0A182QYJ0 A0A182T456 A0A182NLH7 W5JMB5 A0A182VRR2 A0A084W573 A0A182FNS8 A0A182RPB6 T1D515 A0A023EID5 A0A1Q3FBT4 B0W8N3 A0A182JHE6 E1ZVI0 A0A154PPJ6 A0A182JXQ5 F4WYZ2 A0A195BXZ2 A0A195FAS5 A0A158NC85 A0A151WFJ5 A0A0J7L480 A0A0P4WHL6 U5EXH1 A0A0M8ZQA2 E9ILY4 A0A1B6KPZ2 A0A2A3ED51 V9II32 A0A088A7D1 E2BQ20 A0A1B6F1U5 A0A182V8G9 A0A182TJK9 A0A182XHF6 A0A182LG43 Q7PQY7 A0A182HTZ0 A0A0L7R2Z1 A0A195EAB7 A0A026WVX0 D3IWC8 A0A182NZD7 A0A1W4XFG9 A0A097PIE9 A0A067R700 A0A1W4W163 A0A1L8DW98 L0MNA8 B4IKR6 B4R185 P40945 A0A0K8TNS0 A8C9W7 B3MZ91 B4NHH1 B3P9P3 B4L7F7 B4MF16 A0A0N8D229 A0A2H8TP42 Q1W9N5 A0A1Y1N522 Q29CQ9 B4H9Q6 A0A0M4ES39 A0A0P5W4U8 B4PW48 A0A2P2HYT8 A0A0L0C9F7 W8BX92 A0A0K8UWR1 A0A310SM75 A0A2R5LEQ3 A0A293N4R9 E9HDF7 A0A1D2N474 A0A0P6I372 A0A1W7RB47 H9JH97 R7U3Y2 B4JZR6 A0A139WNV7 Q6P5M5 A0A2I9LNP6
Pubmed
23622113
28756777
26354079
22651552
22118469
27649274
+ More
25244985 20920257 23761445 24438588 24330624 24945155 20798317 21719571 21347285 21282665 20966253 12364791 14747013 17210077 24508170 30249741 19837172 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 22936249 8063793 12537569 26369729 17760985 18057021 20479145 28004739 15632085 17550304 26108605 24495485 21292972 27289101 19121390 23254933 18362917 19820115 23594743 29248469
25244985 20920257 23761445 24438588 24330624 24945155 20798317 21719571 21347285 21282665 20966253 12364791 14747013 17210077 24508170 30249741 19837172 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 22936249 8063793 12537569 26369729 17760985 18057021 20479145 28004739 15632085 17550304 26108605 24495485 21292972 27289101 19121390 23254933 18362917 19820115 23594743 29248469
EMBL
GAIX01003955
JAA88605.1
GDQN01001630
JAT89424.1
KZ149894
PZC78882.1
+ More
NWSH01008563 PCG62498.1 ODYU01002049 SOQ39078.1 KQ459989 KPJ18735.1 AK403187 KQ459591 BAM19616.1 KPI96960.1 AGBW02010217 OWR48904.1 BDGG01000002 GAU93133.1 UFQS01001739 UFQT01001739 SSX12131.1 SSX31592.1 GAMD01002784 JAA98806.1 GGFM01001562 MBW22313.1 GGFK01002250 MBW35571.1 AXCM01000497 AXCN02000138 ADMH02001019 GGFL01004310 ETN64433.1 MBW68488.1 ATLV01020512 KE525302 KFB45367.1 GALA01000738 JAA94114.1 GAPW01004913 GAPW01004912 GEHC01000327 JAC08686.1 JAV47318.1 GFDL01010004 JAV25041.1 DS231859 EDS39083.1 GL434534 EFN74817.1 KQ435012 KZC13803.1 GL888463 EGI60646.1 KQ976396 KYM92811.1 KQ981727 KYN37139.1 ADTU01011739 KQ983211 KYQ46602.1 LBMM01000808 KMQ97441.1 GDRN01052471 JAI66298.1 GANO01000958 JAB58913.1 KQ435918 KOX68727.1 GL764129 EFZ18308.1 GEBQ01026462 JAT13515.1 KZ288296 PBC28951.1 JR048856 AEY60783.1 GL449694 EFN82208.1 GECZ01025540 JAS44229.1 AAAB01008859 EAA08117.2 EGK96698.1 APCN01000594 KQ414664 KOC65198.1 KQ979236 KYN22158.1 KK107079 QOIP01000010 EZA60147.1 RLU17371.1 GQ279375 ADB92615.1 KM210090 AIU47295.1 KK852661 KDR19101.1 GFDF01003559 JAV10525.1 AE014135 AGB96577.1 CH480855 EDW52656.1 CM000364 CM002916 EDX13062.1 KMZ07387.1 L25062 AY071450 GDAI01001566 JAI16037.1 AJVK01011187 EU032345 ABV44712.1 CH902633 EDV33696.1 CH964272 EDW83540.1 CH954184 EDV45206.1 CH933813 EDW10951.1 CM000831 CH940665 ACY70521.1 EDW71117.1 GDIP01076415 LRGB01003386 JAM27300.1 KZS02885.1 GFXV01003597 MBW15402.1 ABLF02018887 DQ420640 AK340113 ABD91522.1 BAH70777.1 GEZM01015912 JAV91426.1 CH475429 EAL29264.1 CH479230 EDW36540.1 CP012527 ALC48076.1 GDIP01104813 JAL98901.1 CM000161 EDW99353.1 IACF01001225 LAB66938.1 JRES01000824 KNC28079.1 GAMC01000630 JAC05926.1 GDHF01021202 JAI31112.1 KQ759878 OAD62262.1 GGLE01003856 MBY07982.1 GFWV01022484 MAA47211.1 GL732623 EFX70245.1 LJIJ01000235 ODN00070.1 GDIQ01017805 JAN76932.1 GFAH01000019 JAV48370.1 BABH01012712 AMQN01001788 KB305766 ELU00694.1 CH916380 EDV90932.1 KQ971307 KYB29680.1 CR318588 BC062831 BX571879 AAH62831.1 CAM56630.1 GFWZ01000011 MBW20001.1
NWSH01008563 PCG62498.1 ODYU01002049 SOQ39078.1 KQ459989 KPJ18735.1 AK403187 KQ459591 BAM19616.1 KPI96960.1 AGBW02010217 OWR48904.1 BDGG01000002 GAU93133.1 UFQS01001739 UFQT01001739 SSX12131.1 SSX31592.1 GAMD01002784 JAA98806.1 GGFM01001562 MBW22313.1 GGFK01002250 MBW35571.1 AXCM01000497 AXCN02000138 ADMH02001019 GGFL01004310 ETN64433.1 MBW68488.1 ATLV01020512 KE525302 KFB45367.1 GALA01000738 JAA94114.1 GAPW01004913 GAPW01004912 GEHC01000327 JAC08686.1 JAV47318.1 GFDL01010004 JAV25041.1 DS231859 EDS39083.1 GL434534 EFN74817.1 KQ435012 KZC13803.1 GL888463 EGI60646.1 KQ976396 KYM92811.1 KQ981727 KYN37139.1 ADTU01011739 KQ983211 KYQ46602.1 LBMM01000808 KMQ97441.1 GDRN01052471 JAI66298.1 GANO01000958 JAB58913.1 KQ435918 KOX68727.1 GL764129 EFZ18308.1 GEBQ01026462 JAT13515.1 KZ288296 PBC28951.1 JR048856 AEY60783.1 GL449694 EFN82208.1 GECZ01025540 JAS44229.1 AAAB01008859 EAA08117.2 EGK96698.1 APCN01000594 KQ414664 KOC65198.1 KQ979236 KYN22158.1 KK107079 QOIP01000010 EZA60147.1 RLU17371.1 GQ279375 ADB92615.1 KM210090 AIU47295.1 KK852661 KDR19101.1 GFDF01003559 JAV10525.1 AE014135 AGB96577.1 CH480855 EDW52656.1 CM000364 CM002916 EDX13062.1 KMZ07387.1 L25062 AY071450 GDAI01001566 JAI16037.1 AJVK01011187 EU032345 ABV44712.1 CH902633 EDV33696.1 CH964272 EDW83540.1 CH954184 EDV45206.1 CH933813 EDW10951.1 CM000831 CH940665 ACY70521.1 EDW71117.1 GDIP01076415 LRGB01003386 JAM27300.1 KZS02885.1 GFXV01003597 MBW15402.1 ABLF02018887 DQ420640 AK340113 ABD91522.1 BAH70777.1 GEZM01015912 JAV91426.1 CH475429 EAL29264.1 CH479230 EDW36540.1 CP012527 ALC48076.1 GDIP01104813 JAL98901.1 CM000161 EDW99353.1 IACF01001225 LAB66938.1 JRES01000824 KNC28079.1 GAMC01000630 JAC05926.1 GDHF01021202 JAI31112.1 KQ759878 OAD62262.1 GGLE01003856 MBY07982.1 GFWV01022484 MAA47211.1 GL732623 EFX70245.1 LJIJ01000235 ODN00070.1 GDIQ01017805 JAN76932.1 GFAH01000019 JAV48370.1 BABH01012712 AMQN01001788 KB305766 ELU00694.1 CH916380 EDV90932.1 KQ971307 KYB29680.1 CR318588 BC062831 BX571879 AAH62831.1 CAM56630.1 GFWZ01000011 MBW20001.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000186922
UP000076408
+ More
UP000075883 UP000075886 UP000075901 UP000075884 UP000000673 UP000075920 UP000030765 UP000069272 UP000075900 UP000002320 UP000075880 UP000000311 UP000076502 UP000075881 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000036403 UP000053105 UP000242457 UP000005203 UP000008237 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000053825 UP000078492 UP000053097 UP000279307 UP000075885 UP000192223 UP000027135 UP000192221 UP000000803 UP000001292 UP000000304 UP000092462 UP000007801 UP000007798 UP000008711 UP000009192 UP000008792 UP000076858 UP000007819 UP000001819 UP000008744 UP000092553 UP000002282 UP000037069 UP000000305 UP000094527 UP000005204 UP000014760 UP000001070 UP000007266 UP000000437
UP000075883 UP000075886 UP000075901 UP000075884 UP000000673 UP000075920 UP000030765 UP000069272 UP000075900 UP000002320 UP000075880 UP000000311 UP000076502 UP000075881 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000036403 UP000053105 UP000242457 UP000005203 UP000008237 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000053825 UP000078492 UP000053097 UP000279307 UP000075885 UP000192223 UP000027135 UP000192221 UP000000803 UP000001292 UP000000304 UP000092462 UP000007801 UP000007798 UP000008711 UP000009192 UP000008792 UP000076858 UP000007819 UP000001819 UP000008744 UP000092553 UP000002282 UP000037069 UP000000305 UP000094527 UP000005204 UP000014760 UP000001070 UP000007266 UP000000437
Pfam
PF00025 Arf
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
S4PLM4
A0A1E1WR24
A0A2W1BUX3
A0A2A4ISZ1
A0A2H1VE15
A0A194RN27
+ More
I4DNX6 A0A212F5A4 A0A1D1V306 A0A336L2J2 T1DQ43 A0A182XV13 A0A2M3Z1B8 A0A2M4A499 A0A182M7Z2 A0A182QYJ0 A0A182T456 A0A182NLH7 W5JMB5 A0A182VRR2 A0A084W573 A0A182FNS8 A0A182RPB6 T1D515 A0A023EID5 A0A1Q3FBT4 B0W8N3 A0A182JHE6 E1ZVI0 A0A154PPJ6 A0A182JXQ5 F4WYZ2 A0A195BXZ2 A0A195FAS5 A0A158NC85 A0A151WFJ5 A0A0J7L480 A0A0P4WHL6 U5EXH1 A0A0M8ZQA2 E9ILY4 A0A1B6KPZ2 A0A2A3ED51 V9II32 A0A088A7D1 E2BQ20 A0A1B6F1U5 A0A182V8G9 A0A182TJK9 A0A182XHF6 A0A182LG43 Q7PQY7 A0A182HTZ0 A0A0L7R2Z1 A0A195EAB7 A0A026WVX0 D3IWC8 A0A182NZD7 A0A1W4XFG9 A0A097PIE9 A0A067R700 A0A1W4W163 A0A1L8DW98 L0MNA8 B4IKR6 B4R185 P40945 A0A0K8TNS0 A8C9W7 B3MZ91 B4NHH1 B3P9P3 B4L7F7 B4MF16 A0A0N8D229 A0A2H8TP42 Q1W9N5 A0A1Y1N522 Q29CQ9 B4H9Q6 A0A0M4ES39 A0A0P5W4U8 B4PW48 A0A2P2HYT8 A0A0L0C9F7 W8BX92 A0A0K8UWR1 A0A310SM75 A0A2R5LEQ3 A0A293N4R9 E9HDF7 A0A1D2N474 A0A0P6I372 A0A1W7RB47 H9JH97 R7U3Y2 B4JZR6 A0A139WNV7 Q6P5M5 A0A2I9LNP6
I4DNX6 A0A212F5A4 A0A1D1V306 A0A336L2J2 T1DQ43 A0A182XV13 A0A2M3Z1B8 A0A2M4A499 A0A182M7Z2 A0A182QYJ0 A0A182T456 A0A182NLH7 W5JMB5 A0A182VRR2 A0A084W573 A0A182FNS8 A0A182RPB6 T1D515 A0A023EID5 A0A1Q3FBT4 B0W8N3 A0A182JHE6 E1ZVI0 A0A154PPJ6 A0A182JXQ5 F4WYZ2 A0A195BXZ2 A0A195FAS5 A0A158NC85 A0A151WFJ5 A0A0J7L480 A0A0P4WHL6 U5EXH1 A0A0M8ZQA2 E9ILY4 A0A1B6KPZ2 A0A2A3ED51 V9II32 A0A088A7D1 E2BQ20 A0A1B6F1U5 A0A182V8G9 A0A182TJK9 A0A182XHF6 A0A182LG43 Q7PQY7 A0A182HTZ0 A0A0L7R2Z1 A0A195EAB7 A0A026WVX0 D3IWC8 A0A182NZD7 A0A1W4XFG9 A0A097PIE9 A0A067R700 A0A1W4W163 A0A1L8DW98 L0MNA8 B4IKR6 B4R185 P40945 A0A0K8TNS0 A8C9W7 B3MZ91 B4NHH1 B3P9P3 B4L7F7 B4MF16 A0A0N8D229 A0A2H8TP42 Q1W9N5 A0A1Y1N522 Q29CQ9 B4H9Q6 A0A0M4ES39 A0A0P5W4U8 B4PW48 A0A2P2HYT8 A0A0L0C9F7 W8BX92 A0A0K8UWR1 A0A310SM75 A0A2R5LEQ3 A0A293N4R9 E9HDF7 A0A1D2N474 A0A0P6I372 A0A1W7RB47 H9JH97 R7U3Y2 B4JZR6 A0A139WNV7 Q6P5M5 A0A2I9LNP6
PDB
3AQ4
E-value=6.10078e-80,
Score=753
Ontologies
GO
Topology
Subcellular location
Golgi apparatus
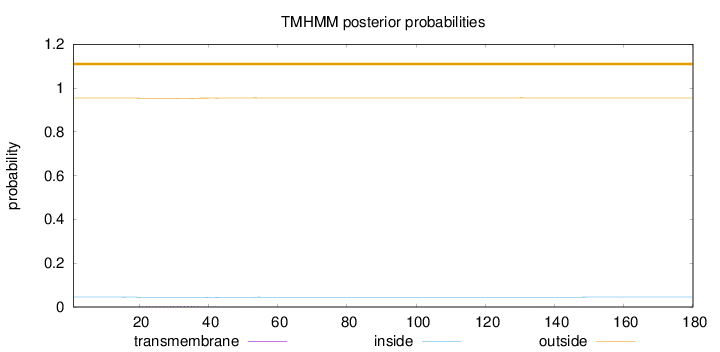
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0901600000000001
Exp number, first 60 AAs:
0.08561
Total prob of N-in:
0.04538
outside
1 - 180
Population Genetic Test Statistics
Pi
282.709377
Theta
210.750621
Tajima's D
0.94764
CLR
0.029228
CSRT
0.64356782160892
Interpretation
Uncertain
