Pre Gene Modal
BGIBMGA008999
Annotation
PREDICTED:_probable_serine/threonine-protein_kinase_nek3_isoform_X4_[Bombyx_mori]
Full name
Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial
+ More
Zinc finger C2HC domain-containing protein 1A
Zinc finger C2HC domain-containing protein 1A
Transcription factor
Location in the cell
Nuclear Reliability : 4.18
Sequence
CDS
ATGCCTTGTCTTGTCTGCGGACGTACTTTTGTGCCACAGTCTCTAGCTAAGCACGTCAAGATTTGCGAGAAGATGACTATTAAAAAACGGAAGACTTTTGATTCCTCAAGACAGAGACGAGAAGGTACAGATTTAGAACAATACTTGCCAAAGAACTTCGGTCTTCCAGAAAACAGTCCATTCCTCGAGAAGAGTCCCCCGCCCACTGCCAAACCCACATTGAAACCTAAACCGCAATCGGTCCGCAGTGCCATAACAAAGCCAATGGCAGAACTGCAGAAGTGTCCGCACTGCGGCCGAGCGTTCGGCGTTCGTGCTTTCGAGAGGCACGTGGAGTGGTGCGCGGACAAGGCCAAGATCTTGCCGGCCGCGCCCGCACACCCGCCGCCGCACATCGCCGACGCCAAGCAGAGACTGAACGCGCGAACACAGTACAAAGCACCCCTGGCTCGAGGTAGAAAAACGTCTCAGTCACGTGACAAATCGTCGAACAGTCGTTCAACGTCTGTAGATTCGTCTCGAGGAGTCTCTCCGCCGCCTCCGCGTGACTGCGGCGACTATGGTGCTGCTAGATCAAGAGCCTCTGGTAATATACACATTCTAAGCTACAAGTATAGATTTAAATCGTTTTAA
Protein
MPCLVCGRTFVPQSLAKHVKICEKMTIKKRKTFDSSRQRREGTDLEQYLPKNFGLPENSPFLEKSPPPTAKPTLKPKPQSVRSAITKPMAELQKCPHCGRAFGVRAFERHVEWCADKAKILPAAPAHPPPHIADAKQRLNARTQYKAPLARGRKTSQSRDKSSNSRSTSVDSSRGVSPPPPRDCGDYGAARSRASGNIHILSYKYRFKSF
Summary
Description
Flavoprotein (FP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
Catalytic Activity
a quinone + succinate = a quinol + fumarate
Cofactor
FAD
Similarity
Belongs to the FAD-dependent oxidoreductase 2 family. FRD/SDH subfamily.
Belongs to the ZC2HC1 family.
Belongs to the ZC2HC1 family.
Keywords
Metal-binding
Zinc
Zinc-finger
Feature
chain Zinc finger C2HC domain-containing protein 1A
Uniprot
A0A2W1C3Y9
A0A1E1WGN1
A0A2H1VE35
A0A0L7LIU5
A0A194PU74
A0A194RRQ4
+ More
A0A182W3S1 A0A182SRD6 A0A182FM27 A0A1B0GKR0 A0A182QM42 A0A1S4HDJ5 A0A182NDK3 A0A182V4R6 A0A182HX86 A0A182WRA0 A0A182JU35 A0A182RBD8 A0A2M4CLF4 A0A084VTQ8 A0A182MS36 A0A182PK86 A0A182JL81 A0A182YLX5 A0A1B6E157 A0A0C9RA44 A0A0C9QJI9 A0A067QQ78 A0A1B6K5C5 A0A336MIL1 A0A336KCQ2 A0A336LYU4 A0A2A3E9N4 A0A139WDG4 A0A139WDK2 A0A2R7WWS9 E9IMU8 A0A0L7QPJ1 A0A0J7L7M0 A0A088A6T2 A0A0A9WBF7 A0A146L0C1 E2B4M4 A0A2S2QE18 E2AFW4 A0A1B6JC03 A0A1B6FMR8 A0A1B6G0I1 A0A195D1B2 J9LI08 J9KNH2 A0A1B6L418 A0A1B6KU29 A0A210QMU4 Q5PPV5 A0A1L8FTC2 A0A131YKW5 E9H118 L7M2A2 A0A099YW95 G1LMV0
A0A182W3S1 A0A182SRD6 A0A182FM27 A0A1B0GKR0 A0A182QM42 A0A1S4HDJ5 A0A182NDK3 A0A182V4R6 A0A182HX86 A0A182WRA0 A0A182JU35 A0A182RBD8 A0A2M4CLF4 A0A084VTQ8 A0A182MS36 A0A182PK86 A0A182JL81 A0A182YLX5 A0A1B6E157 A0A0C9RA44 A0A0C9QJI9 A0A067QQ78 A0A1B6K5C5 A0A336MIL1 A0A336KCQ2 A0A336LYU4 A0A2A3E9N4 A0A139WDG4 A0A139WDK2 A0A2R7WWS9 E9IMU8 A0A0L7QPJ1 A0A0J7L7M0 A0A088A6T2 A0A0A9WBF7 A0A146L0C1 E2B4M4 A0A2S2QE18 E2AFW4 A0A1B6JC03 A0A1B6FMR8 A0A1B6G0I1 A0A195D1B2 J9LI08 J9KNH2 A0A1B6L418 A0A1B6KU29 A0A210QMU4 Q5PPV5 A0A1L8FTC2 A0A131YKW5 E9H118 L7M2A2 A0A099YW95 G1LMV0
EC Number
1.3.5.1
Pubmed
EMBL
KZ149894
PZC78883.1
GDQN01004891
JAT86163.1
ODYU01002049
SOQ39077.1
+ More
JTDY01000907 KOB75483.1 KQ459591 KPI96961.1 KQ459989 KPJ18736.1 AJWK01030933 AJWK01030934 AJWK01030935 AJWK01030936 AJWK01030937 AJWK01030938 AXCN02001758 AAAB01008849 APCN01005154 GGFL01001907 MBW66085.1 ATLV01016450 KE525091 KFB41352.1 AXCM01000169 GEDC01005649 JAS31649.1 GBYB01003686 JAG73453.1 GBYB01003689 JAG73456.1 KK853598 KDR06405.1 GECU01001045 JAT06662.1 UFQS01001264 UFQT01001264 SSX10048.1 SSX29770.1 UFQS01000218 UFQT01000218 SSX01508.1 SSX21888.1 SSX01509.1 SSX21889.1 KZ288312 PBC28435.1 KQ971358 KYB25983.1 KYB25982.1 KK855824 PTY24004.1 GL764255 EFZ18090.1 KQ414819 KOC60474.1 LBMM01000341 KMR00218.1 GBHO01041431 GBHO01041430 GBHO01041429 GDHC01008056 JAG02173.1 JAG02174.1 JAG02175.1 JAQ10573.1 GDHC01017927 JAQ00702.1 GL445574 EFN89377.1 GGMS01006781 MBY75984.1 GL439166 EFN67671.1 GECU01010940 JAS96766.1 GECZ01018273 JAS51496.1 GECZ01013825 JAS55944.1 KQ976973 KYN06700.1 ABLF02020928 ABLF02020932 GEBQ01021592 JAT18385.1 GEBQ01025027 JAT14950.1 NEDP02002776 OWF50052.1 BC087481 CM004477 OCT74830.1 GEDV01008614 JAP79943.1 GL732582 EFX74602.1 GACK01007810 JAA57224.1 KL885833 KGL73223.1 ACTA01100546 ACTA01108546 ACTA01116546 ACTA01124546 ACTA01132546 ACTA01140546 ACTA01148545
JTDY01000907 KOB75483.1 KQ459591 KPI96961.1 KQ459989 KPJ18736.1 AJWK01030933 AJWK01030934 AJWK01030935 AJWK01030936 AJWK01030937 AJWK01030938 AXCN02001758 AAAB01008849 APCN01005154 GGFL01001907 MBW66085.1 ATLV01016450 KE525091 KFB41352.1 AXCM01000169 GEDC01005649 JAS31649.1 GBYB01003686 JAG73453.1 GBYB01003689 JAG73456.1 KK853598 KDR06405.1 GECU01001045 JAT06662.1 UFQS01001264 UFQT01001264 SSX10048.1 SSX29770.1 UFQS01000218 UFQT01000218 SSX01508.1 SSX21888.1 SSX01509.1 SSX21889.1 KZ288312 PBC28435.1 KQ971358 KYB25983.1 KYB25982.1 KK855824 PTY24004.1 GL764255 EFZ18090.1 KQ414819 KOC60474.1 LBMM01000341 KMR00218.1 GBHO01041431 GBHO01041430 GBHO01041429 GDHC01008056 JAG02173.1 JAG02174.1 JAG02175.1 JAQ10573.1 GDHC01017927 JAQ00702.1 GL445574 EFN89377.1 GGMS01006781 MBY75984.1 GL439166 EFN67671.1 GECU01010940 JAS96766.1 GECZ01018273 JAS51496.1 GECZ01013825 JAS55944.1 KQ976973 KYN06700.1 ABLF02020928 ABLF02020932 GEBQ01021592 JAT18385.1 GEBQ01025027 JAT14950.1 NEDP02002776 OWF50052.1 BC087481 CM004477 OCT74830.1 GEDV01008614 JAP79943.1 GL732582 EFX74602.1 GACK01007810 JAA57224.1 KL885833 KGL73223.1 ACTA01100546 ACTA01108546 ACTA01116546 ACTA01124546 ACTA01132546 ACTA01140546 ACTA01148545
Proteomes
UP000037510
UP000053268
UP000053240
UP000075920
UP000075901
UP000069272
+ More
UP000092461 UP000075886 UP000075884 UP000075903 UP000075840 UP000076407 UP000075881 UP000075900 UP000030765 UP000075883 UP000075885 UP000075880 UP000076408 UP000027135 UP000242457 UP000007266 UP000053825 UP000036403 UP000005203 UP000008237 UP000000311 UP000078542 UP000007819 UP000242188 UP000186698 UP000000305 UP000053641 UP000008912
UP000092461 UP000075886 UP000075884 UP000075903 UP000075840 UP000076407 UP000075881 UP000075900 UP000030765 UP000075883 UP000075885 UP000075880 UP000076408 UP000027135 UP000242457 UP000007266 UP000053825 UP000036403 UP000005203 UP000008237 UP000000311 UP000078542 UP000007819 UP000242188 UP000186698 UP000000305 UP000053641 UP000008912
PRIDE
Interpro
IPR026319
ZC2HC1
+ More
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR014748 Enoyl-CoA_hydra_C
IPR026104 ZNF_C2HC_dom_1C
IPR003953 FAD-binding_2
IPR015939 Fum_Rdtase/Succ_DH_flav-like_C
IPR014006 Succ_Dhase_FrdA_Gneg
IPR027477 Succ_DH/fumarate_Rdtase_cat_sf
IPR011281 Succ_DH_flav_su_fwd
IPR036188 FAD/NAD-bd_sf
IPR037099 Fum_R/Succ_DH_flav-like_C_sf
IPR003952 FRD_SDH_FAD_BS
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR014748 Enoyl-CoA_hydra_C
IPR026104 ZNF_C2HC_dom_1C
IPR003953 FAD-binding_2
IPR015939 Fum_Rdtase/Succ_DH_flav-like_C
IPR014006 Succ_Dhase_FrdA_Gneg
IPR027477 Succ_DH/fumarate_Rdtase_cat_sf
IPR011281 Succ_DH_flav_su_fwd
IPR036188 FAD/NAD-bd_sf
IPR037099 Fum_R/Succ_DH_flav-like_C_sf
IPR003952 FRD_SDH_FAD_BS
Gene 3D
ProteinModelPortal
A0A2W1C3Y9
A0A1E1WGN1
A0A2H1VE35
A0A0L7LIU5
A0A194PU74
A0A194RRQ4
+ More
A0A182W3S1 A0A182SRD6 A0A182FM27 A0A1B0GKR0 A0A182QM42 A0A1S4HDJ5 A0A182NDK3 A0A182V4R6 A0A182HX86 A0A182WRA0 A0A182JU35 A0A182RBD8 A0A2M4CLF4 A0A084VTQ8 A0A182MS36 A0A182PK86 A0A182JL81 A0A182YLX5 A0A1B6E157 A0A0C9RA44 A0A0C9QJI9 A0A067QQ78 A0A1B6K5C5 A0A336MIL1 A0A336KCQ2 A0A336LYU4 A0A2A3E9N4 A0A139WDG4 A0A139WDK2 A0A2R7WWS9 E9IMU8 A0A0L7QPJ1 A0A0J7L7M0 A0A088A6T2 A0A0A9WBF7 A0A146L0C1 E2B4M4 A0A2S2QE18 E2AFW4 A0A1B6JC03 A0A1B6FMR8 A0A1B6G0I1 A0A195D1B2 J9LI08 J9KNH2 A0A1B6L418 A0A1B6KU29 A0A210QMU4 Q5PPV5 A0A1L8FTC2 A0A131YKW5 E9H118 L7M2A2 A0A099YW95 G1LMV0
A0A182W3S1 A0A182SRD6 A0A182FM27 A0A1B0GKR0 A0A182QM42 A0A1S4HDJ5 A0A182NDK3 A0A182V4R6 A0A182HX86 A0A182WRA0 A0A182JU35 A0A182RBD8 A0A2M4CLF4 A0A084VTQ8 A0A182MS36 A0A182PK86 A0A182JL81 A0A182YLX5 A0A1B6E157 A0A0C9RA44 A0A0C9QJI9 A0A067QQ78 A0A1B6K5C5 A0A336MIL1 A0A336KCQ2 A0A336LYU4 A0A2A3E9N4 A0A139WDG4 A0A139WDK2 A0A2R7WWS9 E9IMU8 A0A0L7QPJ1 A0A0J7L7M0 A0A088A6T2 A0A0A9WBF7 A0A146L0C1 E2B4M4 A0A2S2QE18 E2AFW4 A0A1B6JC03 A0A1B6FMR8 A0A1B6G0I1 A0A195D1B2 J9LI08 J9KNH2 A0A1B6L418 A0A1B6KU29 A0A210QMU4 Q5PPV5 A0A1L8FTC2 A0A131YKW5 E9H118 L7M2A2 A0A099YW95 G1LMV0
Ontologies
Topology
Subcellular location
Mitochondrion inner membrane
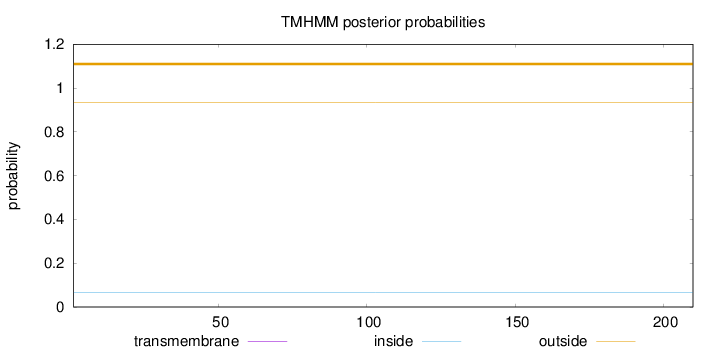
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.06768
outside
1 - 210
Population Genetic Test Statistics
Pi
272.032378
Theta
175.282756
Tajima's D
1.136754
CLR
0.349479
CSRT
0.699265036748163
Interpretation
Uncertain
