Gene
KWMTBOMO01346 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009000
Annotation
PREDICTED:_succinate_dehydrogenase_[ubiquinone]_flavoprotein_subunit?_mitochondrial_[Papilio_xuthus]
Full name
Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial
+ More
Succinate dehydrogenase (quinone)
Succinate dehydrogenase (quinone)
Alternative Name
Flavoprotein subunit of complex II
Succinyl coenzyme A synthetase flavoprotein subunit
Succinyl coenzyme A synthetase flavoprotein subunit
Location in the cell
Mitochondrial Reliability : 2.997
Sequence
CDS
ATGAGTGCATTAATGAAAGCGGCAGTTTCCCGCAGTCCTGCGTCACTGGCATCATACTTATATAGAAATATTCACGTGGCTGCTTCTGGTGCTACTTCTAAAGATGCTTCAGTTACAACAAATATGACTAAAGCATACACAGTAATTGACCACAAACATGATGCATTAGTTATTGGGGCTGGTGGAGCAGGACTTCGAGCAGCCTTTGGTCTTGTCCAAGAAGGATTTAAAACTGCTGTCGTAACTAAGTTGTTCCCTACAAGATCACATACTGTTGCTGCACAGGGTGGAATTAATGCTGCTTTGGGAAACATGGAGGAAGACAGTTGGCTCTGGCATATGTATGACACAGTTAAAGGTTCCGACTGGCTTGGAGATCAGGATGCCATTCACTACATGACTAAAGAAGCGCCTCATGCAGTAATCGAACTGGACAACTACGGAATGCCATTCTCCCGCACCCCGGAGGGAAAGATTTACCAGAGGGCCTTCGGAGGACAGTCTCTCAAGTTCGGAAAGGGAGGACAAGCTCATAGATGCTGCGCCGTAGCTGACAGAACCGGGCACTCCCTCCTTCACACGCTGTACGGCCAATCACTCCGCTACGACTGCGAATACTTCATCGAGTATTTCGCTCTCGATTTGCTAATGGAAGACGGAGTCTGCAAGGGATGCATCGCCATCAATCTCGAAGACGGAACACTGCACAGGTTCCAAGCGAAGAATACGATCTTGGCCACGGGAGGCACCGGCCGCTCATACTTCAGTTGCACGTCGGCGCACACCTGCACAGGGGACGGCACCGCCATGGCCGCTCGAGTCGGTCTGCAGAACGAGGATATGGAGTTTGTGCAGTTCCATCCAACTGGTATCTACGGCGCCGGTTGCCTGATGACCGAGGGTTGTCGTGGCGAGGGCGGCTTCCTGGTCAACTCGAAGGGCGAACGGTTCATGGAACGTTACGCGCCCGTCGCCAAGGACCTGGCCAGTCGAGACGTGGTCTCGCGTGCTATGACGGTTGAAATTATGGAAGGTCGCGGTTGCGGCCCGGAGAAGGACCACGTTCACCTACAGTTGCACCACCTGCCTCCCGAACAACTGAAGCAGCGTCTGCCTGGCATCTCGGAGACGGCCATGATCTTCGCTGGGGTCGACGTCACTAAGGAACCCATTCCTGTGCTACCGACAGTGCACTATAATATGGGTGGTACTCCCACCAATTTCCGCGGGGAGGTGATCACGCACCGTGGCGGCGCGGACCGCGTGGTGGCGGGGCTGCTGGCGGCGGGCGAGGCGTCGTGCGCCAGCGTGCACGGCGCCAACCGGCTGGGCGCCAACTCGCTGCTGGACATCGTGGTGTTCGGCCGCGCCTGCGCACTCACCGTGGCCGACACGGCGCGCCCCGGGGACCCGCAGCCGCCGCTCGCGCCCACAACGGGCGAGGCGAGCATTGCCAACTTGGACAGCATCAGATACGCCAATGGTTCGATCTCTACAGCCGACCTTAGACTTAGAATGCAAAAATGCATGCAGAAGAATGCCGCCGTCTTTAGACAGAAGAGCACCTTGGAGGAAGGTCAACGTCAAATTCACGAGATCTACAAGCAGATAAAAGACGTGAAGGTGTCCGACCGGTCCTTGATCTGGAACAGTGATCTAGTGGAGACGCTGGAGCTTCAGAATCTTCTCATCAACTCGGTACAGATCGTGGAGGGAGCACTGGCGCGGGAGGAGTCGCGCGGAGCCCACGCCCGAGAGGACTTCAAGACCAGACGCGACGAGTACGACTACTCTAAACCTCTCGAGGGACAGAGTAAGTTGCCGTTCGAGCAGCACTGGAGGAAGCACACGCTCGCCGAGACGGACCCCGCGACCGGCGACACGCGCCTCACCTACCGCCCCGTCATCGACCGCACGCTCGACGCCGCCGAGTGCAGCACCGTGCCGCCCGTCATCCGCACCTACTAG
Protein
MSALMKAAVSRSPASLASYLYRNIHVAASGATSKDASVTTNMTKAYTVIDHKHDALVIGAGGAGLRAAFGLVQEGFKTAVVTKLFPTRSHTVAAQGGINAALGNMEEDSWLWHMYDTVKGSDWLGDQDAIHYMTKEAPHAVIELDNYGMPFSRTPEGKIYQRAFGGQSLKFGKGGQAHRCCAVADRTGHSLLHTLYGQSLRYDCEYFIEYFALDLLMEDGVCKGCIAINLEDGTLHRFQAKNTILATGGTGRSYFSCTSAHTCTGDGTAMAARVGLQNEDMEFVQFHPTGIYGAGCLMTEGCRGEGGFLVNSKGERFMERYAPVAKDLASRDVVSRAMTVEIMEGRGCGPEKDHVHLQLHHLPPEQLKQRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGGTPTNFRGEVITHRGGADRVVAGLLAAGEASCASVHGANRLGANSLLDIVVFGRACALTVADTARPGDPQPPLAPTTGEASIANLDSIRYANGSISTADLRLRMQKCMQKNAAVFRQKSTLEEGQRQIHEIYKQIKDVKVSDRSLIWNSDLVETLELQNLLINSVQIVEGALAREESRGAHAREDFKTRRDEYDYSKPLEGQSKLPFEQHWRKHTLAETDPATGDTRLTYRPVIDRTLDAAECSTVPPVIRTY
Summary
Description
Flavoprotein (FP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
Flavoprotein (FP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q). Maintaining electron transport chain function is required to prevent neurodegenerative changes seen in both early- and late-onset disorders.
Flavoprotein (FP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q). Maintaining electron transport chain function is required to prevent neurodegenerative changes seen in both early- and late-onset disorders.
Catalytic Activity
a quinone + succinate = a quinol + fumarate
Cofactor
FAD
Subunit
Component of complex II composed of four subunits: a flavoprotein (FP), an iron-sulfur protein (IP), and a cytochrome b composed of a large and a small subunit.
Similarity
Belongs to the FAD-dependent oxidoreductase 2 family. FRD/SDH subfamily.
Keywords
Complete proteome
Electron transport
FAD
Flavoprotein
Membrane
Mitochondrion
Mitochondrion inner membrane
Oxidoreductase
Reference proteome
Transit peptide
Transport
Tricarboxylic acid cycle
Feature
chain Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial
Uniprot
H9JHK2
A0A2W1BZK9
A0A212FEF5
M9VDW9
A0A1Q3FY45
A0A182KAN5
+ More
A0A1Q3FY46 A0A1Q3FY53 A0A1Q3FY78 A0A182LN92 Q7QDV0 A0A182WR96 A0A182MA54 A0A182NDK7 A0A182ULS7 A0A182HX90 A0A084VTQ4 A0A182YLX1 A0A096ZN66 A0A182W3S5 A0A2M4BGH8 A0A182PVF3 A0A182Q1E1 A0A2M3ZFY9 W5JUV9 A0A182RBD4 A0A182FM31 A0A2M4A5Q6 A0A0A9XPU2 A0A182J2T0 A0A2M4A5S3 A0A2J7Q925 A0A182GZD1 A0A1B6JXD6 U5ELV7 R4WJV3 A0A1B0CLP4 B0WFX7 A0A1L8E0S2 A0A0K2UW91 B4MYF9 A0A0M3QV67 T1E1E5 A0A2D1QW02 B4GDN2 E2AMC7 A0A182TQ59 Q292E4 A0A0J7L7M0 A0A1D2MTD8 A0A1B6E5X9 Q16SE4 A0A3B0J5Z3 A0A1I8Q5A7 T1PAV8 A0A0J9RGM7 B3NJU5 B4PBL8 Q94523 A0A0P5KK93 A0A023EV92 A0A1W4WNT7 B4LJP8 A0A0P5D600 A0A1W4VHE0 A0A336LMK4 A0A0P5AIL3 A0A0P5AEN9 A0A0P5Z0V4 B4JW44 D6WVS3 E2B4M1 A0A3Q0IHL7 A0A0P4WYF9 B4KT93 A0A0P5AMF6 A0A023FCI3 A0A1W0X758 A0A0P4WTD1 A0A0L0BSS9 A0A0P5UMW0 A0A0P5SMH2 A0A0P4WMI9 A0A0P5T1B1 K7ITI9 T1IC04 A0A0P5BVR8 A0A026VYM8 A0A0P6BER3 A0A0P5SY40 F4X086 A0A2I0MWU9 A0A0P5T6T1 B3MGG8 A0A1V4J9E3 A0A0P4ZXE5 A0A099ZDT3 A0A093QLW7 A0A218VA71 H0ZD78
A0A1Q3FY46 A0A1Q3FY53 A0A1Q3FY78 A0A182LN92 Q7QDV0 A0A182WR96 A0A182MA54 A0A182NDK7 A0A182ULS7 A0A182HX90 A0A084VTQ4 A0A182YLX1 A0A096ZN66 A0A182W3S5 A0A2M4BGH8 A0A182PVF3 A0A182Q1E1 A0A2M3ZFY9 W5JUV9 A0A182RBD4 A0A182FM31 A0A2M4A5Q6 A0A0A9XPU2 A0A182J2T0 A0A2M4A5S3 A0A2J7Q925 A0A182GZD1 A0A1B6JXD6 U5ELV7 R4WJV3 A0A1B0CLP4 B0WFX7 A0A1L8E0S2 A0A0K2UW91 B4MYF9 A0A0M3QV67 T1E1E5 A0A2D1QW02 B4GDN2 E2AMC7 A0A182TQ59 Q292E4 A0A0J7L7M0 A0A1D2MTD8 A0A1B6E5X9 Q16SE4 A0A3B0J5Z3 A0A1I8Q5A7 T1PAV8 A0A0J9RGM7 B3NJU5 B4PBL8 Q94523 A0A0P5KK93 A0A023EV92 A0A1W4WNT7 B4LJP8 A0A0P5D600 A0A1W4VHE0 A0A336LMK4 A0A0P5AIL3 A0A0P5AEN9 A0A0P5Z0V4 B4JW44 D6WVS3 E2B4M1 A0A3Q0IHL7 A0A0P4WYF9 B4KT93 A0A0P5AMF6 A0A023FCI3 A0A1W0X758 A0A0P4WTD1 A0A0L0BSS9 A0A0P5UMW0 A0A0P5SMH2 A0A0P4WMI9 A0A0P5T1B1 K7ITI9 T1IC04 A0A0P5BVR8 A0A026VYM8 A0A0P6BER3 A0A0P5SY40 F4X086 A0A2I0MWU9 A0A0P5T6T1 B3MGG8 A0A1V4J9E3 A0A0P4ZXE5 A0A099ZDT3 A0A093QLW7 A0A218VA71 H0ZD78
EC Number
1.3.5.1
Pubmed
19121390
28756777
22118469
20966253
12364791
14747013
+ More
17210077 24438588 25244985 20920257 23761445 25401762 26823975 26483478 23691247 17994087 18057021 24330624 20798317 15632085 23185243 27289101 17510324 25315136 22936249 17550304 10731132 12537572 12537569 10071211 15693940 18599508 24945155 18362917 19820115 25474469 26108605 20075255 24508170 30249741 21719571 23371554 20360741
17210077 24438588 25244985 20920257 23761445 25401762 26823975 26483478 23691247 17994087 18057021 24330624 20798317 15632085 23185243 27289101 17510324 25315136 22936249 17550304 10731132 12537572 12537569 10071211 15693940 18599508 24945155 18362917 19820115 25474469 26108605 20075255 24508170 30249741 21719571 23371554 20360741
EMBL
BABH01012716
BABH01012717
KZ149894
PZC78885.1
AGBW02008935
OWR52136.1
+ More
KC161216 AGJ76544.1 GFDL01002556 JAV32489.1 GFDL01002555 JAV32490.1 GFDL01002551 JAV32494.1 GFDL01002549 JAV32496.1 AAAB01008849 EAA07202.4 AXCM01000168 APCN01005154 ATLV01016449 KE525091 KFB41348.1 KM098145 AIS22434.1 GGFJ01003028 MBW52169.1 AXCN02001756 GGFM01006662 MBW27413.1 ADMH02000002 ETN68137.1 GGFK01002760 MBW36081.1 GBHO01021610 GBRD01008830 GDHC01008054 JAG21994.1 JAG56991.1 JAQ10575.1 GGFK01002780 MBW36101.1 NEVH01016943 PNF25089.1 JXUM01099388 KQ564488 KXJ72205.1 GECU01003842 JAT03865.1 GANO01001255 JAB58616.1 AK417890 BAN21105.1 AJWK01017478 AJWK01017479 AJWK01017480 AJWK01017481 AJWK01017482 AJWK01017483 DS231921 EDS26512.1 GFDF01001754 JAV12330.1 HACA01024595 CDW41956.1 CH963894 EDW77148.1 KRF98587.1 CP012524 ALC41853.1 GALA01001749 JAA93103.1 KY027056 ATP16742.1 CH479181 EDW31689.1 GL440811 EFN65418.1 CM000071 EAL24918.1 KRT01692.1 LBMM01000341 KMR00218.1 LJIJ01000544 ODM96389.1 GEDC01003948 JAS33350.1 CH477675 EAT37381.1 OUUW01000001 SPP75142.1 KA645063 AFP59692.1 CM002911 KMY95086.1 CH954179 EDV55204.1 CM000158 EDW91502.1 AE013599 AY051472 BT099627 Y09064 GDIQ01184056 JAK67669.1 GAPW01000570 JAC13028.1 CH940648 EDW60557.1 KRF79479.1 GDIP01165779 JAJ57623.1 UFQS01000020 UFQT01000020 SSW97497.1 SSX17883.1 GDIP01198727 JAJ24675.1 GDIP01200173 JAJ23229.1 GDIP01128530 GDIP01100377 GDIP01076396 GDIP01051749 GDIP01049839 GDIP01043236 LRGB01000626 JAM51966.1 KZS17456.1 CH916375 EDV98182.1 KQ971358 EFA09226.1 GL445574 EFN89374.1 GDIP01253170 JAI70231.1 CH933808 EDW10605.1 GDIP01196730 JAJ26672.1 GBBI01000219 JAC18493.1 MTYJ01000013 OQV23244.1 GDIP01253413 JAI69988.1 JRES01001422 KNC23038.1 GDIP01110780 JAL92934.1 GDIP01137932 JAL65782.1 GDIP01253314 JAI70087.1 GDIP01139137 GDIP01131546 JAL64577.1 ACPB03004429 GDIP01194428 JAJ28974.1 KK107566 QOIP01000006 EZA48877.1 RLU21947.1 GDIP01015918 JAM87797.1 GDIP01136425 JAL67289.1 GL888493 EGI60151.1 AKCR02000001 PKK34158.1 GDIP01130188 JAL73526.1 CH902619 EDV35711.1 KPU75792.1 LSYS01008581 OPJ68407.1 GDIP01210021 JAJ13381.1 KL891806 KGL79173.1 KL672903 KFW87390.1 MUZQ01000018 OWK62965.1 ABQF01022872
KC161216 AGJ76544.1 GFDL01002556 JAV32489.1 GFDL01002555 JAV32490.1 GFDL01002551 JAV32494.1 GFDL01002549 JAV32496.1 AAAB01008849 EAA07202.4 AXCM01000168 APCN01005154 ATLV01016449 KE525091 KFB41348.1 KM098145 AIS22434.1 GGFJ01003028 MBW52169.1 AXCN02001756 GGFM01006662 MBW27413.1 ADMH02000002 ETN68137.1 GGFK01002760 MBW36081.1 GBHO01021610 GBRD01008830 GDHC01008054 JAG21994.1 JAG56991.1 JAQ10575.1 GGFK01002780 MBW36101.1 NEVH01016943 PNF25089.1 JXUM01099388 KQ564488 KXJ72205.1 GECU01003842 JAT03865.1 GANO01001255 JAB58616.1 AK417890 BAN21105.1 AJWK01017478 AJWK01017479 AJWK01017480 AJWK01017481 AJWK01017482 AJWK01017483 DS231921 EDS26512.1 GFDF01001754 JAV12330.1 HACA01024595 CDW41956.1 CH963894 EDW77148.1 KRF98587.1 CP012524 ALC41853.1 GALA01001749 JAA93103.1 KY027056 ATP16742.1 CH479181 EDW31689.1 GL440811 EFN65418.1 CM000071 EAL24918.1 KRT01692.1 LBMM01000341 KMR00218.1 LJIJ01000544 ODM96389.1 GEDC01003948 JAS33350.1 CH477675 EAT37381.1 OUUW01000001 SPP75142.1 KA645063 AFP59692.1 CM002911 KMY95086.1 CH954179 EDV55204.1 CM000158 EDW91502.1 AE013599 AY051472 BT099627 Y09064 GDIQ01184056 JAK67669.1 GAPW01000570 JAC13028.1 CH940648 EDW60557.1 KRF79479.1 GDIP01165779 JAJ57623.1 UFQS01000020 UFQT01000020 SSW97497.1 SSX17883.1 GDIP01198727 JAJ24675.1 GDIP01200173 JAJ23229.1 GDIP01128530 GDIP01100377 GDIP01076396 GDIP01051749 GDIP01049839 GDIP01043236 LRGB01000626 JAM51966.1 KZS17456.1 CH916375 EDV98182.1 KQ971358 EFA09226.1 GL445574 EFN89374.1 GDIP01253170 JAI70231.1 CH933808 EDW10605.1 GDIP01196730 JAJ26672.1 GBBI01000219 JAC18493.1 MTYJ01000013 OQV23244.1 GDIP01253413 JAI69988.1 JRES01001422 KNC23038.1 GDIP01110780 JAL92934.1 GDIP01137932 JAL65782.1 GDIP01253314 JAI70087.1 GDIP01139137 GDIP01131546 JAL64577.1 ACPB03004429 GDIP01194428 JAJ28974.1 KK107566 QOIP01000006 EZA48877.1 RLU21947.1 GDIP01015918 JAM87797.1 GDIP01136425 JAL67289.1 GL888493 EGI60151.1 AKCR02000001 PKK34158.1 GDIP01130188 JAL73526.1 CH902619 EDV35711.1 KPU75792.1 LSYS01008581 OPJ68407.1 GDIP01210021 JAJ13381.1 KL891806 KGL79173.1 KL672903 KFW87390.1 MUZQ01000018 OWK62965.1 ABQF01022872
Proteomes
UP000005204
UP000007151
UP000075881
UP000075882
UP000007062
UP000076407
+ More
UP000075883 UP000075884 UP000075903 UP000075840 UP000030765 UP000076408 UP000075920 UP000075885 UP000075886 UP000000673 UP000075900 UP000069272 UP000075880 UP000235965 UP000069940 UP000249989 UP000092461 UP000002320 UP000007798 UP000092553 UP000008744 UP000000311 UP000075902 UP000001819 UP000036403 UP000094527 UP000008820 UP000268350 UP000095300 UP000095301 UP000008711 UP000002282 UP000000803 UP000192223 UP000008792 UP000192221 UP000076858 UP000001070 UP000007266 UP000008237 UP000079169 UP000009192 UP000037069 UP000002358 UP000015103 UP000053097 UP000279307 UP000007755 UP000053872 UP000007801 UP000190648 UP000053641 UP000053258 UP000197619 UP000007754
UP000075883 UP000075884 UP000075903 UP000075840 UP000030765 UP000076408 UP000075920 UP000075885 UP000075886 UP000000673 UP000075900 UP000069272 UP000075880 UP000235965 UP000069940 UP000249989 UP000092461 UP000002320 UP000007798 UP000092553 UP000008744 UP000000311 UP000075902 UP000001819 UP000036403 UP000094527 UP000008820 UP000268350 UP000095300 UP000095301 UP000008711 UP000002282 UP000000803 UP000192223 UP000008792 UP000192221 UP000076858 UP000001070 UP000007266 UP000008237 UP000079169 UP000009192 UP000037069 UP000002358 UP000015103 UP000053097 UP000279307 UP000007755 UP000053872 UP000007801 UP000190648 UP000053641 UP000053258 UP000197619 UP000007754
Interpro
Gene 3D
ProteinModelPortal
H9JHK2
A0A2W1BZK9
A0A212FEF5
M9VDW9
A0A1Q3FY45
A0A182KAN5
+ More
A0A1Q3FY46 A0A1Q3FY53 A0A1Q3FY78 A0A182LN92 Q7QDV0 A0A182WR96 A0A182MA54 A0A182NDK7 A0A182ULS7 A0A182HX90 A0A084VTQ4 A0A182YLX1 A0A096ZN66 A0A182W3S5 A0A2M4BGH8 A0A182PVF3 A0A182Q1E1 A0A2M3ZFY9 W5JUV9 A0A182RBD4 A0A182FM31 A0A2M4A5Q6 A0A0A9XPU2 A0A182J2T0 A0A2M4A5S3 A0A2J7Q925 A0A182GZD1 A0A1B6JXD6 U5ELV7 R4WJV3 A0A1B0CLP4 B0WFX7 A0A1L8E0S2 A0A0K2UW91 B4MYF9 A0A0M3QV67 T1E1E5 A0A2D1QW02 B4GDN2 E2AMC7 A0A182TQ59 Q292E4 A0A0J7L7M0 A0A1D2MTD8 A0A1B6E5X9 Q16SE4 A0A3B0J5Z3 A0A1I8Q5A7 T1PAV8 A0A0J9RGM7 B3NJU5 B4PBL8 Q94523 A0A0P5KK93 A0A023EV92 A0A1W4WNT7 B4LJP8 A0A0P5D600 A0A1W4VHE0 A0A336LMK4 A0A0P5AIL3 A0A0P5AEN9 A0A0P5Z0V4 B4JW44 D6WVS3 E2B4M1 A0A3Q0IHL7 A0A0P4WYF9 B4KT93 A0A0P5AMF6 A0A023FCI3 A0A1W0X758 A0A0P4WTD1 A0A0L0BSS9 A0A0P5UMW0 A0A0P5SMH2 A0A0P4WMI9 A0A0P5T1B1 K7ITI9 T1IC04 A0A0P5BVR8 A0A026VYM8 A0A0P6BER3 A0A0P5SY40 F4X086 A0A2I0MWU9 A0A0P5T6T1 B3MGG8 A0A1V4J9E3 A0A0P4ZXE5 A0A099ZDT3 A0A093QLW7 A0A218VA71 H0ZD78
A0A1Q3FY46 A0A1Q3FY53 A0A1Q3FY78 A0A182LN92 Q7QDV0 A0A182WR96 A0A182MA54 A0A182NDK7 A0A182ULS7 A0A182HX90 A0A084VTQ4 A0A182YLX1 A0A096ZN66 A0A182W3S5 A0A2M4BGH8 A0A182PVF3 A0A182Q1E1 A0A2M3ZFY9 W5JUV9 A0A182RBD4 A0A182FM31 A0A2M4A5Q6 A0A0A9XPU2 A0A182J2T0 A0A2M4A5S3 A0A2J7Q925 A0A182GZD1 A0A1B6JXD6 U5ELV7 R4WJV3 A0A1B0CLP4 B0WFX7 A0A1L8E0S2 A0A0K2UW91 B4MYF9 A0A0M3QV67 T1E1E5 A0A2D1QW02 B4GDN2 E2AMC7 A0A182TQ59 Q292E4 A0A0J7L7M0 A0A1D2MTD8 A0A1B6E5X9 Q16SE4 A0A3B0J5Z3 A0A1I8Q5A7 T1PAV8 A0A0J9RGM7 B3NJU5 B4PBL8 Q94523 A0A0P5KK93 A0A023EV92 A0A1W4WNT7 B4LJP8 A0A0P5D600 A0A1W4VHE0 A0A336LMK4 A0A0P5AIL3 A0A0P5AEN9 A0A0P5Z0V4 B4JW44 D6WVS3 E2B4M1 A0A3Q0IHL7 A0A0P4WYF9 B4KT93 A0A0P5AMF6 A0A023FCI3 A0A1W0X758 A0A0P4WTD1 A0A0L0BSS9 A0A0P5UMW0 A0A0P5SMH2 A0A0P4WMI9 A0A0P5T1B1 K7ITI9 T1IC04 A0A0P5BVR8 A0A026VYM8 A0A0P6BER3 A0A0P5SY40 F4X086 A0A2I0MWU9 A0A0P5T6T1 B3MGG8 A0A1V4J9E3 A0A0P4ZXE5 A0A099ZDT3 A0A093QLW7 A0A218VA71 H0ZD78
PDB
2WQY
E-value=0,
Score=2377
Ontologies
PATHWAY
GO
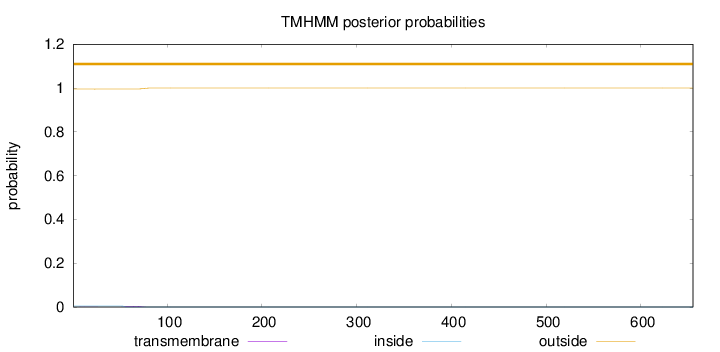
Topology
Subcellular location
Mitochondrion inner membrane
Length:
655
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0871899999999999
Exp number, first 60 AAs:
0.03039
Total prob of N-in:
0.00472
outside
1 - 655
Population Genetic Test Statistics
Pi
206.887721
Theta
172.53391
Tajima's D
1.180786
CLR
0.095668
CSRT
0.713064346782661
Interpretation
Uncertain
