Pre Gene Modal
BGIBMGA008892
Annotation
PREDICTED:_iron-sulfur_cluster_assembly_2_homolog?_mitochondrial_[Amyelois_transitella]
Full name
Iron-sulfur cluster assembly 2 homolog, mitochondrial
Alternative Name
HESB-like domain-containing protein 1
Location in the cell
Nuclear Reliability : 2.266
Sequence
CDS
ATGTTTTTACGTTTTAGAAATTTACACCAGTTAAAAAAGTTTGTTAATGTAAACACATTAAACAAAAGATTCTCCCAAGAACCAGCAACTTCTGTTTCAAGTACAGATATCAGTATTAGTGACAATTGTGTACAAAAATTAAAAGAATTATGTAAAGAAAACGATACCTTTCTGAGGCTTTGTGTAGAAAGTGGTGGATGCTCTGGATTCCAGTATAAATTCAACTTAGACAATGTTGTTGATAGTGATGACAGGATATTTGAAAAAAATGGTGTCAAAGTGGTGGTTGATGAGACGTCACTTGAGTATATTAAAGGTTCAACAATAGATTATCACACTGAACTTATTAGGTCTGCTTTCCGAGTGCAACAGAACCCAAATGCAGATATAGGTTGCTCTTGCGGTGCTAGTTTTTCTATAAAAATTGATTGA
Protein
MFLRFRNLHQLKKFVNVNTLNKRFSQEPATSVSSTDISISDNCVQKLKELCKENDTFLRLCVESGGCSGFQYKFNLDNVVDSDDRIFEKNGVKVVVDETSLEYIKGSTIDYHTELIRSAFRVQQNPNADIGCSCGASFSIKID
Summary
Description
Involved in the maturation of mitochondrial 4Fe-4S proteins functioning late in the iron-sulfur cluster assembly pathway. May be involved in the binding of an intermediate of Fe/S cluster assembly.
Similarity
Belongs to the HesB/IscA family.
Keywords
Alternative splicing
Complete proteome
Disease mutation
Iron
Iron-sulfur
Metal-binding
Mitochondrion
Primary mitochondrial disease
Reference proteome
Transit peptide
Feature
chain Iron-sulfur cluster assembly 2 homolog, mitochondrial
splice variant In isoform 2.
sequence variant In MMDS4; dbSNP:rs730882246.
splice variant In isoform 2.
sequence variant In MMDS4; dbSNP:rs730882246.
Uniprot
H9JH95
A0A212FEH2
A0A2W1C053
A0A2H1WQ37
A0A0P4WKA7
A0A232EWM8
+ More
K7J3W4 A0A0N0U766 A0A0L7R7F6 A0A2T7NYK1 A0A151HZX2 E1ZVH5 A0A2A3ECD0 A0A088AFN0 A0A3M6TF45 A0A195CGG2 F4WYY5 A0A195EBE9 A0A151WFJ3 A0A195F964 A0A2J7PR06 E2BPH3 V4BFF7 A0A026WZ53 J9KAR5 A0A067RJ57 U4UL75 W4ZE95 A0A0J7L919 A0A2H8TF64 A0A2B4SKM6 A0A0C9QV68 J3JYI5 N6T541 A0A139WEV1 A0A1S4EDR9 A0A1X7VN15 A0A2R7W9J4 A0A1S4EDZ6 A0A182HR10 A0A182W8H7 A0A182XH88 Q7Q0Y4 A0A182U397 Q175J2 A0A1S4MWS8 A0A182R1Q2 A0A2S2PFF4 A0A0B6ZSG6 A0A182YQN7 A0A023EG68 A0A182PZS8 B0XLT2 A0A1B6JJ07 A0A182PVP5 A0A182NNS1 A0A0P4VWC1 R4G7R2 A0A1I7Z332 A0A1Y1N8M1 A0A034WQ22 A0A182G0U2 A0A2L1K407 A0A1B6KVV3 A0A2M4C349 A0A1B6EKB2 A0A1D1UI87 A0A1Q3FK48 D4A4L5 A0A1I8MJR8 T1F9C1 A0A0K8UYV4 W5JGC6 F1RBD9 A0A0K8UB87 A0A1J1III5 A0A1I8QBK6 X1WF43 G1KSM7 G3I937 A0A2R8ZSR6 H2R991 A0A2K6CN30 A0A2K6M2I5 A0A2K5U1X3 A0A2K5YIN1 A0A2K6P5Q3 H9EW17 G3SGX5 J9P4R1 Q86U28 A0A2M4AR78 A0A2K5K4R1 A0A096NUJ9 A0A0V0GC83
K7J3W4 A0A0N0U766 A0A0L7R7F6 A0A2T7NYK1 A0A151HZX2 E1ZVH5 A0A2A3ECD0 A0A088AFN0 A0A3M6TF45 A0A195CGG2 F4WYY5 A0A195EBE9 A0A151WFJ3 A0A195F964 A0A2J7PR06 E2BPH3 V4BFF7 A0A026WZ53 J9KAR5 A0A067RJ57 U4UL75 W4ZE95 A0A0J7L919 A0A2H8TF64 A0A2B4SKM6 A0A0C9QV68 J3JYI5 N6T541 A0A139WEV1 A0A1S4EDR9 A0A1X7VN15 A0A2R7W9J4 A0A1S4EDZ6 A0A182HR10 A0A182W8H7 A0A182XH88 Q7Q0Y4 A0A182U397 Q175J2 A0A1S4MWS8 A0A182R1Q2 A0A2S2PFF4 A0A0B6ZSG6 A0A182YQN7 A0A023EG68 A0A182PZS8 B0XLT2 A0A1B6JJ07 A0A182PVP5 A0A182NNS1 A0A0P4VWC1 R4G7R2 A0A1I7Z332 A0A1Y1N8M1 A0A034WQ22 A0A182G0U2 A0A2L1K407 A0A1B6KVV3 A0A2M4C349 A0A1B6EKB2 A0A1D1UI87 A0A1Q3FK48 D4A4L5 A0A1I8MJR8 T1F9C1 A0A0K8UYV4 W5JGC6 F1RBD9 A0A0K8UB87 A0A1J1III5 A0A1I8QBK6 X1WF43 G1KSM7 G3I937 A0A2R8ZSR6 H2R991 A0A2K6CN30 A0A2K6M2I5 A0A2K5U1X3 A0A2K5YIN1 A0A2K6P5Q3 H9EW17 G3SGX5 J9P4R1 Q86U28 A0A2M4AR78 A0A2K5K4R1 A0A096NUJ9 A0A0V0GC83
Pubmed
19121390
22118469
28756777
28648823
20075255
20798317
+ More
30382153 21719571 23254933 24508170 30249741 24845553 23537049 22516182 18362917 19820115 12364791 14747013 17210077 17510324 20566863 25244985 24945155 27129103 26392177 28004739 25348373 26483478 27649274 15057822 15632090 25315136 20920257 23761445 23594743 21804562 22722832 16136131 25362486 17431167 25319552 22398555 16341006 14702039 12508121 15489334 21269460 22323289 23186163 24275569 25539947 25944712
30382153 21719571 23254933 24508170 30249741 24845553 23537049 22516182 18362917 19820115 12364791 14747013 17210077 17510324 20566863 25244985 24945155 27129103 26392177 28004739 25348373 26483478 27649274 15057822 15632090 25315136 20920257 23761445 23594743 21804562 22722832 16136131 25362486 17431167 25319552 22398555 16341006 14702039 12508121 15489334 21269460 22323289 23186163 24275569 25539947 25944712
EMBL
BABH01012718
AGBW02008935
OWR52134.1
KZ149894
PZC78887.1
ODYU01010210
+ More
SOQ55173.1 GDRN01030148 JAI67572.1 NNAY01001838 OXU22754.1 AAZX01005146 KQ435710 KOX79828.1 KQ414642 KOC66774.1 PZQS01000008 PVD26257.1 KQ976642 KYM78691.1 GL434534 EFN74812.1 KZ288301 PBC28859.1 RCHS01003769 RMX39844.1 KQ977873 KYM99173.1 GL888463 EGI60639.1 KQ979236 KYN22164.1 KQ983211 KYQ46597.1 KQ981727 KYN37145.1 NEVH01022633 PNF18771.1 GL449633 EFN82377.1 KB202849 ESO87649.1 KK107061 QOIP01000014 EZA61340.1 RLU14744.1 ABLF02035565 KK852653 KDR19361.1 KDR19363.1 KB632263 ERL90911.1 AAGJ04060801 LBMM01000164 KMR04676.1 GFXV01000914 MBW12719.1 LSMT01000066 PFX29420.1 GBYB01007644 JAG77411.1 BT128313 AEE63271.1 APGK01052223 KB741211 ENN72778.1 KQ971354 KYB26466.1 KK854495 PTY16356.1 APCN01001630 AAAB01008980 EAA14541.4 CH477400 EAT41726.1 EAT41727.1 EAT41728.1 AAZO01006252 GGMR01015551 MBY28170.1 HACG01023835 CEK70700.1 GAPW01005742 JAC07856.1 AXCN02001979 DS234730 EDS34908.1 GECU01008532 JAS99174.1 GDKW01002694 JAI53901.1 ACPB03011086 GAHY01002277 JAA75233.1 GEZM01009840 JAV94272.1 GAKP01003094 JAC55858.1 JXUM01135268 KQ568246 KXJ69085.1 KY827231 AVE16220.1 GEBQ01024404 JAT15573.1 GGFJ01010538 MBW59679.1 GECZ01031405 JAS38364.1 BDGG01000001 GAU89429.1 GFDL01007054 JAV27991.1 AC113727 CH473982 EDL81531.1 AMQM01005305 KB096900 ESO00543.1 GDHF01020543 JAI31771.1 ADMH02001289 ETN63141.1 CR759765 GDHF01028410 JAI23904.1 CVRI01000051 CRK99578.1 JH001565 EGW12998.1 AJFE02085031 AACZ04008420 GABC01010506 GABF01008229 GABE01009231 NBAG03000214 JAA00832.1 JAA13916.1 JAA35508.1 PNI84250.1 AQIA01065493 JSUE03040072 JU322820 JU474273 AFE66576.1 AFH31077.1 CABD030093219 AAEX03005849 BX248252 AK290728 AC005479 CH471061 BC015771 BC032893 GGFK01009901 MBW43222.1 AHZZ02032425 GECL01000458 JAP05666.1
SOQ55173.1 GDRN01030148 JAI67572.1 NNAY01001838 OXU22754.1 AAZX01005146 KQ435710 KOX79828.1 KQ414642 KOC66774.1 PZQS01000008 PVD26257.1 KQ976642 KYM78691.1 GL434534 EFN74812.1 KZ288301 PBC28859.1 RCHS01003769 RMX39844.1 KQ977873 KYM99173.1 GL888463 EGI60639.1 KQ979236 KYN22164.1 KQ983211 KYQ46597.1 KQ981727 KYN37145.1 NEVH01022633 PNF18771.1 GL449633 EFN82377.1 KB202849 ESO87649.1 KK107061 QOIP01000014 EZA61340.1 RLU14744.1 ABLF02035565 KK852653 KDR19361.1 KDR19363.1 KB632263 ERL90911.1 AAGJ04060801 LBMM01000164 KMR04676.1 GFXV01000914 MBW12719.1 LSMT01000066 PFX29420.1 GBYB01007644 JAG77411.1 BT128313 AEE63271.1 APGK01052223 KB741211 ENN72778.1 KQ971354 KYB26466.1 KK854495 PTY16356.1 APCN01001630 AAAB01008980 EAA14541.4 CH477400 EAT41726.1 EAT41727.1 EAT41728.1 AAZO01006252 GGMR01015551 MBY28170.1 HACG01023835 CEK70700.1 GAPW01005742 JAC07856.1 AXCN02001979 DS234730 EDS34908.1 GECU01008532 JAS99174.1 GDKW01002694 JAI53901.1 ACPB03011086 GAHY01002277 JAA75233.1 GEZM01009840 JAV94272.1 GAKP01003094 JAC55858.1 JXUM01135268 KQ568246 KXJ69085.1 KY827231 AVE16220.1 GEBQ01024404 JAT15573.1 GGFJ01010538 MBW59679.1 GECZ01031405 JAS38364.1 BDGG01000001 GAU89429.1 GFDL01007054 JAV27991.1 AC113727 CH473982 EDL81531.1 AMQM01005305 KB096900 ESO00543.1 GDHF01020543 JAI31771.1 ADMH02001289 ETN63141.1 CR759765 GDHF01028410 JAI23904.1 CVRI01000051 CRK99578.1 JH001565 EGW12998.1 AJFE02085031 AACZ04008420 GABC01010506 GABF01008229 GABE01009231 NBAG03000214 JAA00832.1 JAA13916.1 JAA35508.1 PNI84250.1 AQIA01065493 JSUE03040072 JU322820 JU474273 AFE66576.1 AFH31077.1 CABD030093219 AAEX03005849 BX248252 AK290728 AC005479 CH471061 BC015771 BC032893 GGFK01009901 MBW43222.1 AHZZ02032425 GECL01000458 JAP05666.1
Proteomes
UP000005204
UP000007151
UP000215335
UP000002358
UP000053105
UP000053825
+ More
UP000245119 UP000078540 UP000000311 UP000242457 UP000005203 UP000275408 UP000078542 UP000007755 UP000078492 UP000075809 UP000078541 UP000235965 UP000008237 UP000030746 UP000053097 UP000279307 UP000007819 UP000027135 UP000030742 UP000007110 UP000036403 UP000225706 UP000019118 UP000007266 UP000079169 UP000007879 UP000075840 UP000075920 UP000076407 UP000007062 UP000075902 UP000008820 UP000075900 UP000076408 UP000075886 UP000002320 UP000075885 UP000075884 UP000015103 UP000095287 UP000069940 UP000249989 UP000186922 UP000002494 UP000095301 UP000015101 UP000000673 UP000000437 UP000183832 UP000095300 UP000001646 UP000001075 UP000240080 UP000002277 UP000233120 UP000233180 UP000233100 UP000233140 UP000233200 UP000006718 UP000001519 UP000002254 UP000005640 UP000233080 UP000028761
UP000245119 UP000078540 UP000000311 UP000242457 UP000005203 UP000275408 UP000078542 UP000007755 UP000078492 UP000075809 UP000078541 UP000235965 UP000008237 UP000030746 UP000053097 UP000279307 UP000007819 UP000027135 UP000030742 UP000007110 UP000036403 UP000225706 UP000019118 UP000007266 UP000079169 UP000007879 UP000075840 UP000075920 UP000076407 UP000007062 UP000075902 UP000008820 UP000075900 UP000076408 UP000075886 UP000002320 UP000075885 UP000075884 UP000015103 UP000095287 UP000069940 UP000249989 UP000186922 UP000002494 UP000095301 UP000015101 UP000000673 UP000000437 UP000183832 UP000095300 UP000001646 UP000001075 UP000240080 UP000002277 UP000233120 UP000233180 UP000233100 UP000233140 UP000233200 UP000006718 UP000001519 UP000002254 UP000005640 UP000233080 UP000028761
Pfam
PF01521 Fe-S_biosyn
Interpro
SUPFAM
SSF89360
SSF89360
Gene 3D
ProteinModelPortal
H9JH95
A0A212FEH2
A0A2W1C053
A0A2H1WQ37
A0A0P4WKA7
A0A232EWM8
+ More
K7J3W4 A0A0N0U766 A0A0L7R7F6 A0A2T7NYK1 A0A151HZX2 E1ZVH5 A0A2A3ECD0 A0A088AFN0 A0A3M6TF45 A0A195CGG2 F4WYY5 A0A195EBE9 A0A151WFJ3 A0A195F964 A0A2J7PR06 E2BPH3 V4BFF7 A0A026WZ53 J9KAR5 A0A067RJ57 U4UL75 W4ZE95 A0A0J7L919 A0A2H8TF64 A0A2B4SKM6 A0A0C9QV68 J3JYI5 N6T541 A0A139WEV1 A0A1S4EDR9 A0A1X7VN15 A0A2R7W9J4 A0A1S4EDZ6 A0A182HR10 A0A182W8H7 A0A182XH88 Q7Q0Y4 A0A182U397 Q175J2 A0A1S4MWS8 A0A182R1Q2 A0A2S2PFF4 A0A0B6ZSG6 A0A182YQN7 A0A023EG68 A0A182PZS8 B0XLT2 A0A1B6JJ07 A0A182PVP5 A0A182NNS1 A0A0P4VWC1 R4G7R2 A0A1I7Z332 A0A1Y1N8M1 A0A034WQ22 A0A182G0U2 A0A2L1K407 A0A1B6KVV3 A0A2M4C349 A0A1B6EKB2 A0A1D1UI87 A0A1Q3FK48 D4A4L5 A0A1I8MJR8 T1F9C1 A0A0K8UYV4 W5JGC6 F1RBD9 A0A0K8UB87 A0A1J1III5 A0A1I8QBK6 X1WF43 G1KSM7 G3I937 A0A2R8ZSR6 H2R991 A0A2K6CN30 A0A2K6M2I5 A0A2K5U1X3 A0A2K5YIN1 A0A2K6P5Q3 H9EW17 G3SGX5 J9P4R1 Q86U28 A0A2M4AR78 A0A2K5K4R1 A0A096NUJ9 A0A0V0GC83
K7J3W4 A0A0N0U766 A0A0L7R7F6 A0A2T7NYK1 A0A151HZX2 E1ZVH5 A0A2A3ECD0 A0A088AFN0 A0A3M6TF45 A0A195CGG2 F4WYY5 A0A195EBE9 A0A151WFJ3 A0A195F964 A0A2J7PR06 E2BPH3 V4BFF7 A0A026WZ53 J9KAR5 A0A067RJ57 U4UL75 W4ZE95 A0A0J7L919 A0A2H8TF64 A0A2B4SKM6 A0A0C9QV68 J3JYI5 N6T541 A0A139WEV1 A0A1S4EDR9 A0A1X7VN15 A0A2R7W9J4 A0A1S4EDZ6 A0A182HR10 A0A182W8H7 A0A182XH88 Q7Q0Y4 A0A182U397 Q175J2 A0A1S4MWS8 A0A182R1Q2 A0A2S2PFF4 A0A0B6ZSG6 A0A182YQN7 A0A023EG68 A0A182PZS8 B0XLT2 A0A1B6JJ07 A0A182PVP5 A0A182NNS1 A0A0P4VWC1 R4G7R2 A0A1I7Z332 A0A1Y1N8M1 A0A034WQ22 A0A182G0U2 A0A2L1K407 A0A1B6KVV3 A0A2M4C349 A0A1B6EKB2 A0A1D1UI87 A0A1Q3FK48 D4A4L5 A0A1I8MJR8 T1F9C1 A0A0K8UYV4 W5JGC6 F1RBD9 A0A0K8UB87 A0A1J1III5 A0A1I8QBK6 X1WF43 G1KSM7 G3I937 A0A2R8ZSR6 H2R991 A0A2K6CN30 A0A2K6M2I5 A0A2K5U1X3 A0A2K5YIN1 A0A2K6P5Q3 H9EW17 G3SGX5 J9P4R1 Q86U28 A0A2M4AR78 A0A2K5K4R1 A0A096NUJ9 A0A0V0GC83
PDB
1NWB
E-value=7.86232e-18,
Score=215
Ontologies
GO
Topology
Subcellular location
Mitochondrion
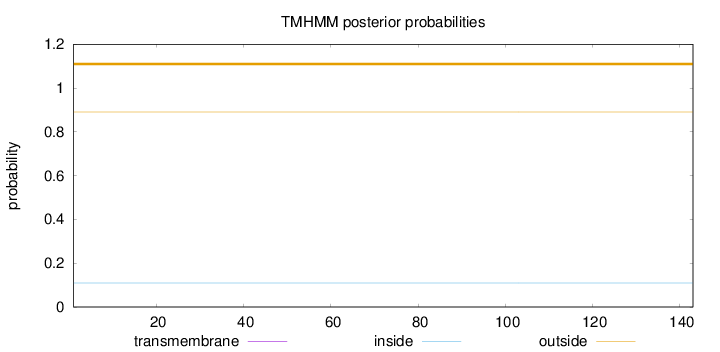
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10992
outside
1 - 143
Population Genetic Test Statistics
Pi
198.350245
Theta
187.137756
Tajima's D
0.35125
CLR
0.323666
CSRT
0.464726763661817
Interpretation
Uncertain
