Gene
KWMTBOMO01340
Pre Gene Modal
BGIBMGA009003
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_split_ends_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.362 Nuclear Reliability : 1.353
Sequence
CDS
ATGTCTGCAGTATACGGAGGCGCGCGGTCCCCGCCCCCCGCGCACGGCCGCCCCCTCGCCGCCCCGCCGGGCCCGCCGCACGCCAGCCAGGTGCCGCGGGAGGCTGACTCCTTGCAGATGCTCCTTAGGAGATACCCGGTGATGTGGCAAGGTCTGTTGGCTCTGAAGAACGACTCTGCGGCCGTACAGATGCACTTCGTGGGCGGTAACCCTGGCGTGGCGGGCGACACGCTGTCTCGGCACCACGAGCAGGCCACGCCGCTGCTCCGTATCGCTCAGCGCATGCGCCTCGAGCCCGCGCAGCTGGACCAGGTCCACCGCAAGATGAAGATGGAAAACGAACATTGTATGTTGCTCGCATTACCATGCGGACGAGATCATATGGACGTCTTGCAGCAGTCTAACAACCTTACTGCTGGTTTTATAACGTACTTGCAACGTAAACAAGCCGCTGGAATTGTTAATATCGCTCCACCTGGCCATCATCAGGCAATGTTTACCGTTCACATATTTCCTTCATGCGATTTCGCAAACGAAAACCTGAATCGCATAGCGCCCGACTTGATGCATCGTGTCGCCGACATTGCGCATTTGTTAATTGTCATCGCAACTGTGTGA
Protein
MSAVYGGARSPPPAHGRPLAAPPGPPHASQVPREADSLQMLLRRYPVMWQGLLALKNDSAAVQMHFVGGNPGVAGDTLSRHHEQATPLLRIAQRMRLEPAQLDQVHRKMKMENEHCMLLALPCGRDHMDVLQQSNNLTAGFITYLQRKQAAGIVNIAPPGHHQAMFTVHIFPSCDFANENLNRIAPDLMHRVADIAHLLIVIATV
Summary
Uniprot
H9JHK5
A0A2H1WGY7
A0A1E1WFY3
A0A2W1C186
A0A194RMJ1
A0A212FH59
+ More
A0A194PUG8 A0A1B0CA31 A0A1B6KAU7 A0A2A3E9K8 A0A1B0DR41 A0A1B0REL4 A0A182KZ42 A0A0A9WBB1 A0A182G8W6 A0A1B6DNN1 A0A1B6D8R2 A0A1B6E9Y0 A0A1B6D7H4 A0A1Y1MNY4 A0A088A6Q3 A0A2J7Q937 A0A2J7Q932 K7IRP5 A0A232ESN1 A0A0M9A9M3 E0VE07 A0A182G6I8 A0A1B6FUY1 A0A067R5I5 A0A182Y6A2 B0W710 A0A182F557 T1IFC0 Q16VU7 A0A1S4FMM6 W5J954 E9GPR5 W4VRA4 A0A182UZJ7 A0A182T4Q2 A0A084VSR8 A0A182J2U4 A0A1B6G8L0 A0A1Y9GKD0 A0A293M5E4 T1J4C3 A0A336LLB9 A0A310SCZ7 A0A293MAL5 A0A0K8VFZ0 A0A034VZY0 A0A224YW08 A0A2P8Y518 A0A0J7L6Z3 E2A976 E2BUB2 A0A1Z5L7K2 A0A182Q6X9 A0A2R7W5Z9 A0A2R7W6I8 A0A0K8WIZ4 A0A034W511 A0A0K8U5C2 A0A3L8DCL0 A0A224XHC0 A0A026WS22 Q1WWE0 A0A182LW95 A0A182RWR0 A0A182W8R4 A0A0T6BAV3 A0A195B697 A0A147BCR4 A0A131XZ99 A0A182NP23 A0A182JW56 A0A195C3K3 A0A1I8NIP2 A0A1I8NIP3 D6WWB2 A0A151XJZ9 A0A0P6HFX0 A0A182P7Z7 T1PA64 A0A164KPC2 A0A131Z7P5 A0A131YJB0 A0A0P5LW22 A0A195EKP7 A0A195FSC1 A7UV52 F4WCD7 A0A158NRE2 A0A0P5AH96 A0A0P5K6B8 A0A0P5JPT8 A0A0P4ZQK9 A0A0P6HAP2 A0A0N8AU67
A0A194PUG8 A0A1B0CA31 A0A1B6KAU7 A0A2A3E9K8 A0A1B0DR41 A0A1B0REL4 A0A182KZ42 A0A0A9WBB1 A0A182G8W6 A0A1B6DNN1 A0A1B6D8R2 A0A1B6E9Y0 A0A1B6D7H4 A0A1Y1MNY4 A0A088A6Q3 A0A2J7Q937 A0A2J7Q932 K7IRP5 A0A232ESN1 A0A0M9A9M3 E0VE07 A0A182G6I8 A0A1B6FUY1 A0A067R5I5 A0A182Y6A2 B0W710 A0A182F557 T1IFC0 Q16VU7 A0A1S4FMM6 W5J954 E9GPR5 W4VRA4 A0A182UZJ7 A0A182T4Q2 A0A084VSR8 A0A182J2U4 A0A1B6G8L0 A0A1Y9GKD0 A0A293M5E4 T1J4C3 A0A336LLB9 A0A310SCZ7 A0A293MAL5 A0A0K8VFZ0 A0A034VZY0 A0A224YW08 A0A2P8Y518 A0A0J7L6Z3 E2A976 E2BUB2 A0A1Z5L7K2 A0A182Q6X9 A0A2R7W5Z9 A0A2R7W6I8 A0A0K8WIZ4 A0A034W511 A0A0K8U5C2 A0A3L8DCL0 A0A224XHC0 A0A026WS22 Q1WWE0 A0A182LW95 A0A182RWR0 A0A182W8R4 A0A0T6BAV3 A0A195B697 A0A147BCR4 A0A131XZ99 A0A182NP23 A0A182JW56 A0A195C3K3 A0A1I8NIP2 A0A1I8NIP3 D6WWB2 A0A151XJZ9 A0A0P6HFX0 A0A182P7Z7 T1PA64 A0A164KPC2 A0A131Z7P5 A0A131YJB0 A0A0P5LW22 A0A195EKP7 A0A195FSC1 A7UV52 F4WCD7 A0A158NRE2 A0A0P5AH96 A0A0P5K6B8 A0A0P5JPT8 A0A0P4ZQK9 A0A0P6HAP2 A0A0N8AU67
Pubmed
19121390
28756777
26354079
22118469
20966253
25401762
+ More
26483478 28004739 20075255 28648823 20566863 24845553 25244985 17510324 20920257 23761445 21292972 24438588 25348373 28797301 29403074 20798317 28528879 30249741 24508170 29652888 25315136 18362917 19820115 26830274 12364791 21719571 21347285
26483478 28004739 20075255 28648823 20566863 24845553 25244985 17510324 20920257 23761445 21292972 24438588 25348373 28797301 29403074 20798317 28528879 30249741 24508170 29652888 25315136 18362917 19820115 26830274 12364791 21719571 21347285
EMBL
BABH01012733
BABH01012734
BABH01012735
BABH01012736
ODYU01008601
SOQ52329.1
+ More
GDQN01005134 JAT85920.1 KZ149894 PZC78890.1 KQ459989 KPJ18742.1 AGBW02008559 OWR53043.1 KQ459591 KPI96967.1 AJWK01003195 AJWK01003196 GEBQ01031663 JAT08314.1 KZ288312 PBC28408.1 AJVK01008905 AJVK01008906 AJVK01008907 KM264383 AKE98363.1 GBHO01039791 JAG03813.1 JXUM01048580 JXUM01048581 KQ561568 KXJ78134.1 GEDC01010012 JAS27286.1 GEDC01015246 JAS22052.1 GEDC01002634 JAS34664.1 GEDC01015667 JAS21631.1 GEZM01027268 JAV86788.1 NEVH01016943 PNF25099.1 PNF25100.1 NNAY01002394 OXU21365.1 KQ435709 KOX80030.1 DS235088 EEB11613.1 JXUM01007573 JXUM01007574 KQ560243 KXJ83624.1 GECZ01015761 JAS54008.1 KK852871 KDR14584.1 DS231851 EDS37284.1 ACPB03008176 ACPB03008177 CH477580 EAT38712.1 ADMH02002089 ETN59395.1 GL732557 EFX78544.1 GANO01003950 JAB55921.1 ATLV01016139 ATLV01016140 KE525057 KFB41012.1 GECZ01010993 JAS58776.1 APCN01001659 APCN01001660 GFWV01010539 MAA35268.1 JH431842 UFQS01000003 UFQT01000003 SSW96760.1 SSX17147.1 KQ776753 OAD52187.1 GFWV01013264 MAA37993.1 GDHF01028338 GDHF01020690 GDHF01020066 GDHF01014561 JAI23976.1 JAI31624.1 JAI32248.1 JAI37753.1 GAKP01010096 JAC48856.1 GFPF01009969 MAA21115.1 PYGN01000923 PSN39346.1 LBMM01000455 KMQ98299.1 GL437711 EFN70061.1 GL450619 EFN80740.1 GFJQ02003577 JAW03393.1 AXCN02001964 KK854317 PTY14581.1 PTY14580.1 GDHF01001252 JAI51062.1 GAKP01010095 GAKP01010094 JAC48858.1 GDHF01030452 JAI21862.1 QOIP01000009 RLU18215.1 GFTR01008995 JAW07431.1 KK107119 EZA58827.1 BT024966 ABE01196.1 AXCM01000709 LJIG01002470 KRT84470.1 KQ976582 KYM79780.1 GEGO01006853 JAR88551.1 GEFM01004208 JAP71588.1 KQ978292 KYM95444.1 KQ971361 EFA08686.2 KQ982052 KYQ60661.1 GDIQ01029197 JAN65540.1 KA645030 AFP59659.1 LRGB01003275 KZS03435.1 GEDV01001178 JAP87379.1 GEDV01009932 JAP78625.1 GDIQ01164545 JAK87180.1 KQ978739 KYN28813.1 KQ981305 KYN42799.1 AAAB01008980 EDO63446.1 GL888070 EGI68117.1 ADTU01001951 GDIP01198859 JAJ24543.1 GDIQ01188535 JAK63190.1 GDIQ01196685 GDIQ01096666 JAK55040.1 GDIP01213157 JAJ10245.1 GDIQ01031546 JAN63191.1 GDIQ01242523 JAK09202.1
GDQN01005134 JAT85920.1 KZ149894 PZC78890.1 KQ459989 KPJ18742.1 AGBW02008559 OWR53043.1 KQ459591 KPI96967.1 AJWK01003195 AJWK01003196 GEBQ01031663 JAT08314.1 KZ288312 PBC28408.1 AJVK01008905 AJVK01008906 AJVK01008907 KM264383 AKE98363.1 GBHO01039791 JAG03813.1 JXUM01048580 JXUM01048581 KQ561568 KXJ78134.1 GEDC01010012 JAS27286.1 GEDC01015246 JAS22052.1 GEDC01002634 JAS34664.1 GEDC01015667 JAS21631.1 GEZM01027268 JAV86788.1 NEVH01016943 PNF25099.1 PNF25100.1 NNAY01002394 OXU21365.1 KQ435709 KOX80030.1 DS235088 EEB11613.1 JXUM01007573 JXUM01007574 KQ560243 KXJ83624.1 GECZ01015761 JAS54008.1 KK852871 KDR14584.1 DS231851 EDS37284.1 ACPB03008176 ACPB03008177 CH477580 EAT38712.1 ADMH02002089 ETN59395.1 GL732557 EFX78544.1 GANO01003950 JAB55921.1 ATLV01016139 ATLV01016140 KE525057 KFB41012.1 GECZ01010993 JAS58776.1 APCN01001659 APCN01001660 GFWV01010539 MAA35268.1 JH431842 UFQS01000003 UFQT01000003 SSW96760.1 SSX17147.1 KQ776753 OAD52187.1 GFWV01013264 MAA37993.1 GDHF01028338 GDHF01020690 GDHF01020066 GDHF01014561 JAI23976.1 JAI31624.1 JAI32248.1 JAI37753.1 GAKP01010096 JAC48856.1 GFPF01009969 MAA21115.1 PYGN01000923 PSN39346.1 LBMM01000455 KMQ98299.1 GL437711 EFN70061.1 GL450619 EFN80740.1 GFJQ02003577 JAW03393.1 AXCN02001964 KK854317 PTY14581.1 PTY14580.1 GDHF01001252 JAI51062.1 GAKP01010095 GAKP01010094 JAC48858.1 GDHF01030452 JAI21862.1 QOIP01000009 RLU18215.1 GFTR01008995 JAW07431.1 KK107119 EZA58827.1 BT024966 ABE01196.1 AXCM01000709 LJIG01002470 KRT84470.1 KQ976582 KYM79780.1 GEGO01006853 JAR88551.1 GEFM01004208 JAP71588.1 KQ978292 KYM95444.1 KQ971361 EFA08686.2 KQ982052 KYQ60661.1 GDIQ01029197 JAN65540.1 KA645030 AFP59659.1 LRGB01003275 KZS03435.1 GEDV01001178 JAP87379.1 GEDV01009932 JAP78625.1 GDIQ01164545 JAK87180.1 KQ978739 KYN28813.1 KQ981305 KYN42799.1 AAAB01008980 EDO63446.1 GL888070 EGI68117.1 ADTU01001951 GDIP01198859 JAJ24543.1 GDIQ01188535 JAK63190.1 GDIQ01196685 GDIQ01096666 JAK55040.1 GDIP01213157 JAJ10245.1 GDIQ01031546 JAN63191.1 GDIQ01242523 JAK09202.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000092461
UP000242457
+ More
UP000092462 UP000075882 UP000069940 UP000249989 UP000005203 UP000235965 UP000002358 UP000215335 UP000053105 UP000009046 UP000027135 UP000076408 UP000002320 UP000069272 UP000015103 UP000008820 UP000000673 UP000000305 UP000075903 UP000075901 UP000030765 UP000075880 UP000075840 UP000245037 UP000036403 UP000000311 UP000008237 UP000075886 UP000279307 UP000053097 UP000075883 UP000075900 UP000075920 UP000078540 UP000075884 UP000075881 UP000078542 UP000095301 UP000007266 UP000075809 UP000075885 UP000076858 UP000078492 UP000078541 UP000007062 UP000007755 UP000005205
UP000092462 UP000075882 UP000069940 UP000249989 UP000005203 UP000235965 UP000002358 UP000215335 UP000053105 UP000009046 UP000027135 UP000076408 UP000002320 UP000069272 UP000015103 UP000008820 UP000000673 UP000000305 UP000075903 UP000075901 UP000030765 UP000075880 UP000075840 UP000245037 UP000036403 UP000000311 UP000008237 UP000075886 UP000279307 UP000053097 UP000075883 UP000075900 UP000075920 UP000078540 UP000075884 UP000075881 UP000078542 UP000095301 UP000007266 UP000075809 UP000075885 UP000076858 UP000078492 UP000078541 UP000007062 UP000007755 UP000005205
Interpro
IPR012921
SPOC_C
+ More
IPR035979 RBD_domain_sf
IPR016194 SPOC-like_C_dom_sf
IPR010912 SPOC_met
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR034174 SHARP_RRM3
IPR000504 RRM_dom
IPR034175 SHARP_RRM4
IPR034173 SHARP_RRM2
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR034172 SHARP_RRM1
IPR035899 DBL_dom_sf
IPR000219 DH-domain
IPR011993 PH-like_dom_sf
IPR035979 RBD_domain_sf
IPR016194 SPOC-like_C_dom_sf
IPR010912 SPOC_met
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR034174 SHARP_RRM3
IPR000504 RRM_dom
IPR034175 SHARP_RRM4
IPR034173 SHARP_RRM2
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR034172 SHARP_RRM1
IPR035899 DBL_dom_sf
IPR000219 DH-domain
IPR011993 PH-like_dom_sf
Gene 3D
ProteinModelPortal
H9JHK5
A0A2H1WGY7
A0A1E1WFY3
A0A2W1C186
A0A194RMJ1
A0A212FH59
+ More
A0A194PUG8 A0A1B0CA31 A0A1B6KAU7 A0A2A3E9K8 A0A1B0DR41 A0A1B0REL4 A0A182KZ42 A0A0A9WBB1 A0A182G8W6 A0A1B6DNN1 A0A1B6D8R2 A0A1B6E9Y0 A0A1B6D7H4 A0A1Y1MNY4 A0A088A6Q3 A0A2J7Q937 A0A2J7Q932 K7IRP5 A0A232ESN1 A0A0M9A9M3 E0VE07 A0A182G6I8 A0A1B6FUY1 A0A067R5I5 A0A182Y6A2 B0W710 A0A182F557 T1IFC0 Q16VU7 A0A1S4FMM6 W5J954 E9GPR5 W4VRA4 A0A182UZJ7 A0A182T4Q2 A0A084VSR8 A0A182J2U4 A0A1B6G8L0 A0A1Y9GKD0 A0A293M5E4 T1J4C3 A0A336LLB9 A0A310SCZ7 A0A293MAL5 A0A0K8VFZ0 A0A034VZY0 A0A224YW08 A0A2P8Y518 A0A0J7L6Z3 E2A976 E2BUB2 A0A1Z5L7K2 A0A182Q6X9 A0A2R7W5Z9 A0A2R7W6I8 A0A0K8WIZ4 A0A034W511 A0A0K8U5C2 A0A3L8DCL0 A0A224XHC0 A0A026WS22 Q1WWE0 A0A182LW95 A0A182RWR0 A0A182W8R4 A0A0T6BAV3 A0A195B697 A0A147BCR4 A0A131XZ99 A0A182NP23 A0A182JW56 A0A195C3K3 A0A1I8NIP2 A0A1I8NIP3 D6WWB2 A0A151XJZ9 A0A0P6HFX0 A0A182P7Z7 T1PA64 A0A164KPC2 A0A131Z7P5 A0A131YJB0 A0A0P5LW22 A0A195EKP7 A0A195FSC1 A7UV52 F4WCD7 A0A158NRE2 A0A0P5AH96 A0A0P5K6B8 A0A0P5JPT8 A0A0P4ZQK9 A0A0P6HAP2 A0A0N8AU67
A0A194PUG8 A0A1B0CA31 A0A1B6KAU7 A0A2A3E9K8 A0A1B0DR41 A0A1B0REL4 A0A182KZ42 A0A0A9WBB1 A0A182G8W6 A0A1B6DNN1 A0A1B6D8R2 A0A1B6E9Y0 A0A1B6D7H4 A0A1Y1MNY4 A0A088A6Q3 A0A2J7Q937 A0A2J7Q932 K7IRP5 A0A232ESN1 A0A0M9A9M3 E0VE07 A0A182G6I8 A0A1B6FUY1 A0A067R5I5 A0A182Y6A2 B0W710 A0A182F557 T1IFC0 Q16VU7 A0A1S4FMM6 W5J954 E9GPR5 W4VRA4 A0A182UZJ7 A0A182T4Q2 A0A084VSR8 A0A182J2U4 A0A1B6G8L0 A0A1Y9GKD0 A0A293M5E4 T1J4C3 A0A336LLB9 A0A310SCZ7 A0A293MAL5 A0A0K8VFZ0 A0A034VZY0 A0A224YW08 A0A2P8Y518 A0A0J7L6Z3 E2A976 E2BUB2 A0A1Z5L7K2 A0A182Q6X9 A0A2R7W5Z9 A0A2R7W6I8 A0A0K8WIZ4 A0A034W511 A0A0K8U5C2 A0A3L8DCL0 A0A224XHC0 A0A026WS22 Q1WWE0 A0A182LW95 A0A182RWR0 A0A182W8R4 A0A0T6BAV3 A0A195B697 A0A147BCR4 A0A131XZ99 A0A182NP23 A0A182JW56 A0A195C3K3 A0A1I8NIP2 A0A1I8NIP3 D6WWB2 A0A151XJZ9 A0A0P6HFX0 A0A182P7Z7 T1PA64 A0A164KPC2 A0A131Z7P5 A0A131YJB0 A0A0P5LW22 A0A195EKP7 A0A195FSC1 A7UV52 F4WCD7 A0A158NRE2 A0A0P5AH96 A0A0P5K6B8 A0A0P5JPT8 A0A0P4ZQK9 A0A0P6HAP2 A0A0N8AU67
PDB
2RT5
E-value=5.01565e-46,
Score=461
Ontologies
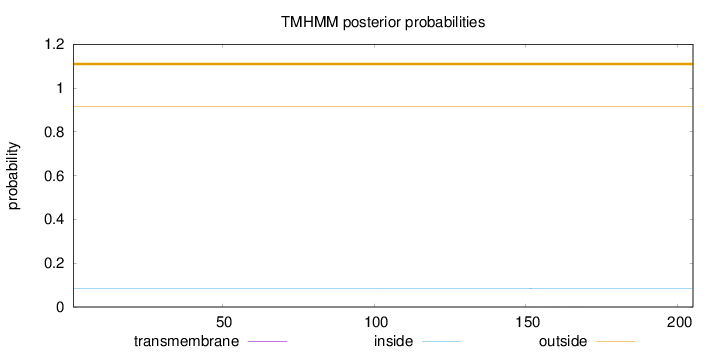
Topology
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01365
Exp number, first 60 AAs:
0.00105
Total prob of N-in:
0.08568
outside
1 - 205
Population Genetic Test Statistics
Pi
308.881169
Theta
231.362472
Tajima's D
1.219997
CLR
0
CSRT
0.715214239288036
Interpretation
Uncertain
