Gene
KWMTBOMO01336
Pre Gene Modal
BGIBMGA008888
Annotation
Uncharacterized_protein_OBRU01_13176_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.388 Mitochondrial Reliability : 1.088 Nuclear Reliability : 1.341
Sequence
CDS
ATGTTCCCAGAAGAAGGGTGGGCAAGAAGCGCATCTTCTTCTTACTGGACGTTGCAGCCCTGTTGGTGGCGTCGATCGCGTTGCAAGGTAGTCGAAGTCGCAGGCACTAGAAGGCACAGCACCCACGCTCGGATGGTCGTGAGCGGAGTCAATGCTGTTTATATCGTGGGAACATTTAAGCATCTCGGAACCGACGCTGATTTTAAGTTATACCTGACAACTAACGTGACCCAGGCCGACTTCAACATGGGCTACACGATGACCGGGACATTAGAACGTGGAAGTCGTTCTGCCAACTCCTTCCAAGTGACACACTTTGCTGTATTGCGCCGTTGCGACCACGACACGCACCATTTGAAAAACTCCTAA
Protein
MFPEEGWARSASSSYWTLQPCWWRRSRCKVVEVAGTRRHSTHARMVVSGVNAVYIVGTFKHLGTDADFKLYLTTNVTQADFNMGYTMTGTLERGSRSANSFQVTHFAVLRRCDHDTHHLKNS
Summary
Uniprot
A0A2H1VC12
A0A2W1C1Q6
A0A0L7KYJ6
A0A212FH31
A0A067RGJ1
E2B2C7
+ More
E2AJF3 F4WNB8 A0A195FEB9 V9IKE8 A0A087ZW01 A0A0L7R536 A0A310SQ26 A0A158NG91 A0A151XAI5 A0A151J5A8 A0A232EZ71 K7J3F9 A0A2A4JWI0 A0A1S4EC63 A0A182QLZ6 A0A2Y9D0T1 A0A1S4H8G5 A0A182UYE1 A0A3L8DME4 A0A2S2Q3D3 J9M6U8 A0A2S2NGS3 A0A0P5EJG0 A0A164NIN2 A0A0P6AFN5 A0A210PTT2 K1PQC1 A0A1A9UWK9 V4B1X5 K1PCZ5 A0A087UT42 A0A1A9X4A6 B4MRD0 A0A1B0A2Q6 A0A1A9YF64 B4KMH7 Q5TMK8 E9GTB0 B4J4S8 A0A182J400 A0A1B0FE47 A0A1B0BNR6 B4LK44 A0A1I8M5W4 A0A0L0CFH2 A0A0P6I9W3 A0A2T7Q0J7 R7TFG4 B4QBW6 B4PAW5 Q9W194 B4I2F4 B3NQH0 A0A0J9RJV1 A0A1I8PX55 B5DZ81 A0A1W4UMP5 B4H698 A0A0R3UJV6 A0A1W0WHK9 A0A195B743 E9IMQ1 A0A068XT53 A0A0R3T0B1 A0A0R3SJM0 A0A0R3X2V4 A0A3P7L5V3 A0A183T502 A0A0R3W0M8 A0A068Y4J8
E2AJF3 F4WNB8 A0A195FEB9 V9IKE8 A0A087ZW01 A0A0L7R536 A0A310SQ26 A0A158NG91 A0A151XAI5 A0A151J5A8 A0A232EZ71 K7J3F9 A0A2A4JWI0 A0A1S4EC63 A0A182QLZ6 A0A2Y9D0T1 A0A1S4H8G5 A0A182UYE1 A0A3L8DME4 A0A2S2Q3D3 J9M6U8 A0A2S2NGS3 A0A0P5EJG0 A0A164NIN2 A0A0P6AFN5 A0A210PTT2 K1PQC1 A0A1A9UWK9 V4B1X5 K1PCZ5 A0A087UT42 A0A1A9X4A6 B4MRD0 A0A1B0A2Q6 A0A1A9YF64 B4KMH7 Q5TMK8 E9GTB0 B4J4S8 A0A182J400 A0A1B0FE47 A0A1B0BNR6 B4LK44 A0A1I8M5W4 A0A0L0CFH2 A0A0P6I9W3 A0A2T7Q0J7 R7TFG4 B4QBW6 B4PAW5 Q9W194 B4I2F4 B3NQH0 A0A0J9RJV1 A0A1I8PX55 B5DZ81 A0A1W4UMP5 B4H698 A0A0R3UJV6 A0A1W0WHK9 A0A195B743 E9IMQ1 A0A068XT53 A0A0R3T0B1 A0A0R3SJM0 A0A0R3X2V4 A0A3P7L5V3 A0A183T502 A0A0R3W0M8 A0A068Y4J8
Pubmed
28756777
26227816
22118469
24845553
20798317
21719571
+ More
21347285 28648823 20075255 12364791 30249741 28812685 22992520 23254933 17994087 21292972 25315136 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 21282665 23485966
21347285 28648823 20075255 12364791 30249741 28812685 22992520 23254933 17994087 21292972 25315136 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 21282665 23485966
EMBL
ODYU01001753
SOQ38388.1
KZ149894
PZC78896.1
JTDY01004285
KOB68338.1
+ More
AGBW02008559 OWR53038.1 KK852699 KDR18229.1 GL445130 EFN90164.1 GL439972 EFN66474.1 GL888236 EGI64288.1 KQ981625 KYN39025.1 JR049927 AEY61127.1 KQ414654 KOC65939.1 KQ761783 OAD57015.1 ADTU01015059 ADTU01015060 ADTU01015061 ADTU01015062 ADTU01015063 KQ982339 KYQ57376.1 KQ980017 KYN18218.1 NNAY01001530 OXU23671.1 NWSH01000444 PCG76375.1 AXCN02001651 APCN01002379 AAAB01008986 QOIP01000006 RLU21486.1 GGMS01003035 MBY72238.1 ABLF02021074 ABLF02021076 ABLF02021079 ABLF02021080 GGMR01003751 GGMR01012520 MBY16370.1 MBY25139.1 GDIQ01268895 JAJ82829.1 LRGB01002851 KZS05992.1 GDIP01030218 JAM73497.1 NEDP02005500 OWF39893.1 JH816959 EKC23873.1 KB200521 ESP01391.1 JH816085 EKC21692.1 KK121470 KFM80531.1 CH963850 EDW74669.1 CH933808 EDW10824.2 EAL38889.3 GL732563 EFX77391.1 CH916367 EDW00624.1 CCAG010020544 JXJN01017575 CH940648 EDW60633.1 JRES01000470 KNC30990.1 GDIQ01024209 JAN70528.1 PZQS01000001 PVD39193.1 AMQN01013260 KB310083 ELT92518.1 CM000362 EDX08521.1 CM000158 EDW92505.1 AE013599 BT010276 AAF47183.2 AAQ23594.1 CH480820 EDW53949.1 CH954179 EDV56973.1 CM002911 KMY96293.1 CM000071 EDY69552.1 CH479213 EDW33322.1 UXSR01005413 VDD81817.1 MTYJ01000100 OQV14691.1 KQ976574 KYM80338.1 GL764255 EFZ18143.1 UZAE01000051 VDN96094.1 UYSG01002381 VDL57451.1 UYWX01020389 VDM32044.1 UYRU01054796 VDN12805.1 UYSU01036619 VDL97935.1 UYRS01018292 VDK31470.1 LN902843 CDS37110.1
AGBW02008559 OWR53038.1 KK852699 KDR18229.1 GL445130 EFN90164.1 GL439972 EFN66474.1 GL888236 EGI64288.1 KQ981625 KYN39025.1 JR049927 AEY61127.1 KQ414654 KOC65939.1 KQ761783 OAD57015.1 ADTU01015059 ADTU01015060 ADTU01015061 ADTU01015062 ADTU01015063 KQ982339 KYQ57376.1 KQ980017 KYN18218.1 NNAY01001530 OXU23671.1 NWSH01000444 PCG76375.1 AXCN02001651 APCN01002379 AAAB01008986 QOIP01000006 RLU21486.1 GGMS01003035 MBY72238.1 ABLF02021074 ABLF02021076 ABLF02021079 ABLF02021080 GGMR01003751 GGMR01012520 MBY16370.1 MBY25139.1 GDIQ01268895 JAJ82829.1 LRGB01002851 KZS05992.1 GDIP01030218 JAM73497.1 NEDP02005500 OWF39893.1 JH816959 EKC23873.1 KB200521 ESP01391.1 JH816085 EKC21692.1 KK121470 KFM80531.1 CH963850 EDW74669.1 CH933808 EDW10824.2 EAL38889.3 GL732563 EFX77391.1 CH916367 EDW00624.1 CCAG010020544 JXJN01017575 CH940648 EDW60633.1 JRES01000470 KNC30990.1 GDIQ01024209 JAN70528.1 PZQS01000001 PVD39193.1 AMQN01013260 KB310083 ELT92518.1 CM000362 EDX08521.1 CM000158 EDW92505.1 AE013599 BT010276 AAF47183.2 AAQ23594.1 CH480820 EDW53949.1 CH954179 EDV56973.1 CM002911 KMY96293.1 CM000071 EDY69552.1 CH479213 EDW33322.1 UXSR01005413 VDD81817.1 MTYJ01000100 OQV14691.1 KQ976574 KYM80338.1 GL764255 EFZ18143.1 UZAE01000051 VDN96094.1 UYSG01002381 VDL57451.1 UYWX01020389 VDM32044.1 UYRU01054796 VDN12805.1 UYSU01036619 VDL97935.1 UYRS01018292 VDK31470.1 LN902843 CDS37110.1
Proteomes
UP000037510
UP000007151
UP000027135
UP000008237
UP000000311
UP000007755
+ More
UP000078541 UP000005203 UP000053825 UP000005205 UP000075809 UP000078492 UP000215335 UP000002358 UP000218220 UP000079169 UP000075886 UP000075840 UP000075903 UP000279307 UP000007819 UP000076858 UP000242188 UP000005408 UP000078200 UP000030746 UP000054359 UP000091820 UP000007798 UP000092445 UP000092443 UP000009192 UP000007062 UP000000305 UP000001070 UP000075880 UP000092444 UP000092460 UP000008792 UP000095301 UP000037069 UP000245119 UP000014760 UP000000304 UP000002282 UP000000803 UP000001292 UP000008711 UP000095300 UP000001819 UP000192221 UP000008744 UP000046399 UP000267029 UP000078540 UP000046398 UP000278807 UP000046397 UP000274504 UP000046396 UP000274429 UP000050788 UP000275846 UP000046400 UP000282613 UP000017246
UP000078541 UP000005203 UP000053825 UP000005205 UP000075809 UP000078492 UP000215335 UP000002358 UP000218220 UP000079169 UP000075886 UP000075840 UP000075903 UP000279307 UP000007819 UP000076858 UP000242188 UP000005408 UP000078200 UP000030746 UP000054359 UP000091820 UP000007798 UP000092445 UP000092443 UP000009192 UP000007062 UP000000305 UP000001070 UP000075880 UP000092444 UP000092460 UP000008792 UP000095301 UP000037069 UP000245119 UP000014760 UP000000304 UP000002282 UP000000803 UP000001292 UP000008711 UP000095300 UP000001819 UP000192221 UP000008744 UP000046399 UP000267029 UP000078540 UP000046398 UP000278807 UP000046397 UP000274504 UP000046396 UP000274429 UP000050788 UP000275846 UP000046400 UP000282613 UP000017246
PRIDE
Interpro
SUPFAM
SSF57756
SSF57756
ProteinModelPortal
A0A2H1VC12
A0A2W1C1Q6
A0A0L7KYJ6
A0A212FH31
A0A067RGJ1
E2B2C7
+ More
E2AJF3 F4WNB8 A0A195FEB9 V9IKE8 A0A087ZW01 A0A0L7R536 A0A310SQ26 A0A158NG91 A0A151XAI5 A0A151J5A8 A0A232EZ71 K7J3F9 A0A2A4JWI0 A0A1S4EC63 A0A182QLZ6 A0A2Y9D0T1 A0A1S4H8G5 A0A182UYE1 A0A3L8DME4 A0A2S2Q3D3 J9M6U8 A0A2S2NGS3 A0A0P5EJG0 A0A164NIN2 A0A0P6AFN5 A0A210PTT2 K1PQC1 A0A1A9UWK9 V4B1X5 K1PCZ5 A0A087UT42 A0A1A9X4A6 B4MRD0 A0A1B0A2Q6 A0A1A9YF64 B4KMH7 Q5TMK8 E9GTB0 B4J4S8 A0A182J400 A0A1B0FE47 A0A1B0BNR6 B4LK44 A0A1I8M5W4 A0A0L0CFH2 A0A0P6I9W3 A0A2T7Q0J7 R7TFG4 B4QBW6 B4PAW5 Q9W194 B4I2F4 B3NQH0 A0A0J9RJV1 A0A1I8PX55 B5DZ81 A0A1W4UMP5 B4H698 A0A0R3UJV6 A0A1W0WHK9 A0A195B743 E9IMQ1 A0A068XT53 A0A0R3T0B1 A0A0R3SJM0 A0A0R3X2V4 A0A3P7L5V3 A0A183T502 A0A0R3W0M8 A0A068Y4J8
E2AJF3 F4WNB8 A0A195FEB9 V9IKE8 A0A087ZW01 A0A0L7R536 A0A310SQ26 A0A158NG91 A0A151XAI5 A0A151J5A8 A0A232EZ71 K7J3F9 A0A2A4JWI0 A0A1S4EC63 A0A182QLZ6 A0A2Y9D0T1 A0A1S4H8G5 A0A182UYE1 A0A3L8DME4 A0A2S2Q3D3 J9M6U8 A0A2S2NGS3 A0A0P5EJG0 A0A164NIN2 A0A0P6AFN5 A0A210PTT2 K1PQC1 A0A1A9UWK9 V4B1X5 K1PCZ5 A0A087UT42 A0A1A9X4A6 B4MRD0 A0A1B0A2Q6 A0A1A9YF64 B4KMH7 Q5TMK8 E9GTB0 B4J4S8 A0A182J400 A0A1B0FE47 A0A1B0BNR6 B4LK44 A0A1I8M5W4 A0A0L0CFH2 A0A0P6I9W3 A0A2T7Q0J7 R7TFG4 B4QBW6 B4PAW5 Q9W194 B4I2F4 B3NQH0 A0A0J9RJV1 A0A1I8PX55 B5DZ81 A0A1W4UMP5 B4H698 A0A0R3UJV6 A0A1W0WHK9 A0A195B743 E9IMQ1 A0A068XT53 A0A0R3T0B1 A0A0R3SJM0 A0A0R3X2V4 A0A3P7L5V3 A0A183T502 A0A0R3W0M8 A0A068Y4J8
Ontologies
GO
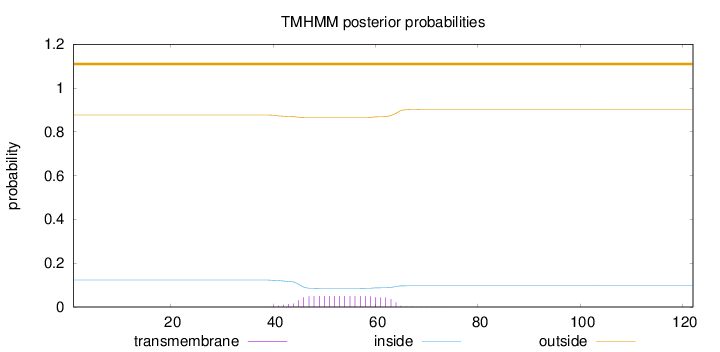
Topology
Length:
122
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.9648
Exp number, first 60 AAs:
0.80826
Total prob of N-in:
0.12307
outside
1 - 122
Population Genetic Test Statistics
Pi
228.760325
Theta
170.677583
Tajima's D
0.934253
CLR
0.195532
CSRT
0.646517674116294
Interpretation
Uncertain
