Gene
KWMTBOMO01335 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008887
Annotation
glutaredoxin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.897 Nuclear Reliability : 1.297
Sequence
CDS
ATGGGTTCTCAGTCTGGAAAAATTACGCGTTCATCTAAAATGGCCGGATCTATAGACATTCAACAGTTTATCAAGGAAGCTATCTCCAAAGACAAAGTTGTAGTGTTCTCCAAATCTTACTGTCCTTACTGTAAGCTGGCAAAAGATGTTTTTGAGAAAGTGAAGCAACCAATTAAAGTTATTGAGTTGAATGAACGTGATGATGGAAACACCATTCAAGATAATCTCGCACAACTGACTGGTTTCAGAACTGTACCTCAAGTCTTTATAAATGGCAACTGTGTGGGAGGTGGCTCTGATGTTAAAGCATTGTATGAATCTGGAAAATTAGAACCTATGTTAATAGGATAA
Protein
MGSQSGKITRSSKMAGSIDIQQFIKEAISKDKVVVFSKSYCPYCKLAKDVFEKVKQPIKVIELNERDDGNTIQDNLAQLTGFRTVPQVFINGNCVGGGSDVKALYESGKLEPMLIG
Summary
Uniprot
Q2F5R3
A0A2H1WWC3
A0A1L2D9B5
H9JH90
A0A2A4JX73
I4DKA9
+ More
S4PWY8 A0A194Q0N9 A0A212FH36 R4WCS3 A0A2R7VZZ6 A0A069DP08 A0A0N7Z8W8 R4FME4 A0A023F874 A0A0K8SAU0 A0A0A9XTG8 A0A0A9XNM0 A0A0K8S3W8 A0A0T6ATB1 A0A1B6GJ49 H2M9V6 A0A3Q0IJ42 A0A1S3CTP4 A0A3P9H2F3 Q1HQT9 A0A1S4E6Y6 A0A1B6LAX0 A0A3P9K4W8 J9HGS4 A0A3P8ZFE4 A0A1B6D7Z4 A0A1Q3FDY3 A0A182QAI6 E0VX45 A0A1B6DU45 G3NQZ4 A0A3B3C1Z1 Q16HL7 B0WPR4 G3NQZ9 A0A0K8TNB9 T1D5F9 Q4S6P0 C3KJX2 A0A182N254 A0A1S3KIX1 A0A1S3KIH6 V5GAP1 A0A2P8ZNQ0 H3D465 A0A060X233 A0A1S3KIH5 A0A1B6EM83 A0A1A7YW09 A0A2U9C1Q3 A0A195DN31 A0A1A8I8S2 A0A1A8AWY1 A0A1A8MD12 A0A3B4VF81 A0A182P5E7 A0A3B4VG43 A0A0J7KWM5 J3JX82 A0A2Y9D1B8 A0A3F2YZM4 A0A182V2F2 A0A182X5T2 Q7QHA7 A0A182HW63 A0A023EF22 A0A2U9C4A2 A0A1A8CL62 A0A1A8AEU7 N6STT7 A0A1A8HR78 A0A1B6GN29 A0A1Y1MPP9 A0A0S7M3S0 A0A3B4Y0V3 A0A182WDP7 E2A1C0 A0A1Y1MRM2 A0A1B6GUB1 I3JY28 A0A182YAX1 A0A3B4EQ68 A0A3P8R5Z7 A0A3P9CFQ2 A0A1L8DU31 A0A3P8UHR5 A0A1W4X8G7 A0A182R6M3 A0A084W8B9 A0A3B4Z9H0 A0A3P8R654 A0A3F2YVC4
S4PWY8 A0A194Q0N9 A0A212FH36 R4WCS3 A0A2R7VZZ6 A0A069DP08 A0A0N7Z8W8 R4FME4 A0A023F874 A0A0K8SAU0 A0A0A9XTG8 A0A0A9XNM0 A0A0K8S3W8 A0A0T6ATB1 A0A1B6GJ49 H2M9V6 A0A3Q0IJ42 A0A1S3CTP4 A0A3P9H2F3 Q1HQT9 A0A1S4E6Y6 A0A1B6LAX0 A0A3P9K4W8 J9HGS4 A0A3P8ZFE4 A0A1B6D7Z4 A0A1Q3FDY3 A0A182QAI6 E0VX45 A0A1B6DU45 G3NQZ4 A0A3B3C1Z1 Q16HL7 B0WPR4 G3NQZ9 A0A0K8TNB9 T1D5F9 Q4S6P0 C3KJX2 A0A182N254 A0A1S3KIX1 A0A1S3KIH6 V5GAP1 A0A2P8ZNQ0 H3D465 A0A060X233 A0A1S3KIH5 A0A1B6EM83 A0A1A7YW09 A0A2U9C1Q3 A0A195DN31 A0A1A8I8S2 A0A1A8AWY1 A0A1A8MD12 A0A3B4VF81 A0A182P5E7 A0A3B4VG43 A0A0J7KWM5 J3JX82 A0A2Y9D1B8 A0A3F2YZM4 A0A182V2F2 A0A182X5T2 Q7QHA7 A0A182HW63 A0A023EF22 A0A2U9C4A2 A0A1A8CL62 A0A1A8AEU7 N6STT7 A0A1A8HR78 A0A1B6GN29 A0A1Y1MPP9 A0A0S7M3S0 A0A3B4Y0V3 A0A182WDP7 E2A1C0 A0A1Y1MRM2 A0A1B6GUB1 I3JY28 A0A182YAX1 A0A3B4EQ68 A0A3P8R5Z7 A0A3P9CFQ2 A0A1L8DU31 A0A3P8UHR5 A0A1W4X8G7 A0A182R6M3 A0A084W8B9 A0A3B4Z9H0 A0A3P8R654 A0A3F2YVC4
Pubmed
28756777
19121390
22651552
23622113
26354079
22118469
+ More
23691247 26334808 27129103 25474469 25401762 26823975 17554307 17204158 17510324 25069045 20566863 29451363 26369729 24330624 15496914 29403074 24755649 22516182 20966253 12364791 14747013 17210077 24945155 23537049 28004739 20798317 25186727 25244985 24487278 24438588
23691247 26334808 27129103 25474469 25401762 26823975 17554307 17204158 17510324 25069045 20566863 29451363 26369729 24330624 15496914 29403074 24755649 22516182 20966253 12364791 14747013 17210077 24945155 23537049 28004739 20798317 25186727 25244985 24487278 24438588
EMBL
DQ311360
ABD36304.1
ODYU01011499
SOQ57266.1
KU356683
KZ149894
+ More
AMW64458.1 PZC78897.1 BABH01012748 BABH01012749 NWSH01000444 PCG76376.1 AK401727 BAM18349.1 GAIX01005963 JAA86597.1 KQ459591 KPI96975.1 AGBW02008559 OWR53037.1 AK417123 BAN20338.1 KK854206 PTY13067.1 GBGD01003513 JAC85376.1 GDKW01002187 JAI54408.1 ACPB03016562 GAHY01001744 JAA75766.1 GBBI01001191 JAC17521.1 GBRD01017853 GBRD01015956 JAG49870.1 GBHO01023180 GBHO01023179 GDHC01015076 GDHC01001917 JAG20424.1 JAG20425.1 JAQ03553.1 JAQ16712.1 GBHO01023181 GBHO01002495 GBRD01017852 GBRD01017849 JAG20423.1 JAG41109.1 JAG47975.1 GBRD01017851 GBRD01015960 GDHC01011813 GDHC01009440 JAG47976.1 JAQ06816.1 JAQ09189.1 LJIG01022855 KRT78371.1 GECZ01007303 JAS62466.1 DQ440355 ABF18388.1 GEBQ01019127 JAT20850.1 CH478160 EJY58072.1 GEDC01015506 JAS21792.1 GFDL01009284 JAV25761.1 AXCN02001743 DS235829 EEB17951.1 GEDC01008098 JAS29200.1 EAT33741.1 DS232028 EDS32473.1 GDAI01001734 JAI15869.1 GALA01000528 JAA94324.1 CAAE01014724 CAG03692.1 BT083237 ACQ58944.1 GALX01001276 JAB67190.1 PYGN01000008 PSN58141.1 FR904890 CDQ73267.1 GECZ01030737 GECZ01023622 JAS39032.1 JAS46147.1 HADX01012560 SBP34792.1 CP026254 AWP10555.1 KQ980713 KYN14283.1 HAED01007285 SBQ93497.1 HADY01021192 HAEJ01002991 SBP59677.1 HAEF01013543 HAEG01012421 SBR54702.1 LBMM01002569 KMQ94649.1 BT127850 AEE62812.1 AAAB01008816 EAA05108.2 APCN01001469 GAPW01006228 JAC07370.1 AWP10556.1 HADZ01016533 SBP80474.1 HADY01014491 SBP52976.1 APGK01056608 KB741277 KB631611 ENN71079.1 ERL84649.1 HAED01000747 SBQ86592.1 GECZ01005952 JAS63817.1 GEZM01029795 GEZM01029794 JAV85926.1 GBYX01119920 JAO96480.1 GL435760 EFN72746.1 GEZM01029793 JAV85927.1 GECZ01003768 GECZ01001197 JAS66001.1 JAS68572.1 AERX01031367 GFDF01004148 JAV09936.1 ATLV01021410 KE525318 KFB46463.1 AXCM01001638
AMW64458.1 PZC78897.1 BABH01012748 BABH01012749 NWSH01000444 PCG76376.1 AK401727 BAM18349.1 GAIX01005963 JAA86597.1 KQ459591 KPI96975.1 AGBW02008559 OWR53037.1 AK417123 BAN20338.1 KK854206 PTY13067.1 GBGD01003513 JAC85376.1 GDKW01002187 JAI54408.1 ACPB03016562 GAHY01001744 JAA75766.1 GBBI01001191 JAC17521.1 GBRD01017853 GBRD01015956 JAG49870.1 GBHO01023180 GBHO01023179 GDHC01015076 GDHC01001917 JAG20424.1 JAG20425.1 JAQ03553.1 JAQ16712.1 GBHO01023181 GBHO01002495 GBRD01017852 GBRD01017849 JAG20423.1 JAG41109.1 JAG47975.1 GBRD01017851 GBRD01015960 GDHC01011813 GDHC01009440 JAG47976.1 JAQ06816.1 JAQ09189.1 LJIG01022855 KRT78371.1 GECZ01007303 JAS62466.1 DQ440355 ABF18388.1 GEBQ01019127 JAT20850.1 CH478160 EJY58072.1 GEDC01015506 JAS21792.1 GFDL01009284 JAV25761.1 AXCN02001743 DS235829 EEB17951.1 GEDC01008098 JAS29200.1 EAT33741.1 DS232028 EDS32473.1 GDAI01001734 JAI15869.1 GALA01000528 JAA94324.1 CAAE01014724 CAG03692.1 BT083237 ACQ58944.1 GALX01001276 JAB67190.1 PYGN01000008 PSN58141.1 FR904890 CDQ73267.1 GECZ01030737 GECZ01023622 JAS39032.1 JAS46147.1 HADX01012560 SBP34792.1 CP026254 AWP10555.1 KQ980713 KYN14283.1 HAED01007285 SBQ93497.1 HADY01021192 HAEJ01002991 SBP59677.1 HAEF01013543 HAEG01012421 SBR54702.1 LBMM01002569 KMQ94649.1 BT127850 AEE62812.1 AAAB01008816 EAA05108.2 APCN01001469 GAPW01006228 JAC07370.1 AWP10556.1 HADZ01016533 SBP80474.1 HADY01014491 SBP52976.1 APGK01056608 KB741277 KB631611 ENN71079.1 ERL84649.1 HAED01000747 SBQ86592.1 GECZ01005952 JAS63817.1 GEZM01029795 GEZM01029794 JAV85926.1 GBYX01119920 JAO96480.1 GL435760 EFN72746.1 GEZM01029793 JAV85927.1 GECZ01003768 GECZ01001197 JAS66001.1 JAS68572.1 AERX01031367 GFDF01004148 JAV09936.1 ATLV01021410 KE525318 KFB46463.1 AXCM01001638
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000015103
UP000001038
+ More
UP000079169 UP000265200 UP000265180 UP000008820 UP000265140 UP000075886 UP000009046 UP000007635 UP000261560 UP000002320 UP000075884 UP000087266 UP000245037 UP000007303 UP000193380 UP000246464 UP000078492 UP000261420 UP000075885 UP000036403 UP000075882 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000019118 UP000030742 UP000261360 UP000075920 UP000000311 UP000005207 UP000076408 UP000261460 UP000265100 UP000265160 UP000265120 UP000192223 UP000075900 UP000030765 UP000261400 UP000075883
UP000079169 UP000265200 UP000265180 UP000008820 UP000265140 UP000075886 UP000009046 UP000007635 UP000261560 UP000002320 UP000075884 UP000087266 UP000245037 UP000007303 UP000193380 UP000246464 UP000078492 UP000261420 UP000075885 UP000036403 UP000075882 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000019118 UP000030742 UP000261360 UP000075920 UP000000311 UP000005207 UP000076408 UP000261460 UP000265100 UP000265160 UP000265120 UP000192223 UP000075900 UP000030765 UP000261400 UP000075883
Pfam
PF00462 Glutaredoxin
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
Q2F5R3
A0A2H1WWC3
A0A1L2D9B5
H9JH90
A0A2A4JX73
I4DKA9
+ More
S4PWY8 A0A194Q0N9 A0A212FH36 R4WCS3 A0A2R7VZZ6 A0A069DP08 A0A0N7Z8W8 R4FME4 A0A023F874 A0A0K8SAU0 A0A0A9XTG8 A0A0A9XNM0 A0A0K8S3W8 A0A0T6ATB1 A0A1B6GJ49 H2M9V6 A0A3Q0IJ42 A0A1S3CTP4 A0A3P9H2F3 Q1HQT9 A0A1S4E6Y6 A0A1B6LAX0 A0A3P9K4W8 J9HGS4 A0A3P8ZFE4 A0A1B6D7Z4 A0A1Q3FDY3 A0A182QAI6 E0VX45 A0A1B6DU45 G3NQZ4 A0A3B3C1Z1 Q16HL7 B0WPR4 G3NQZ9 A0A0K8TNB9 T1D5F9 Q4S6P0 C3KJX2 A0A182N254 A0A1S3KIX1 A0A1S3KIH6 V5GAP1 A0A2P8ZNQ0 H3D465 A0A060X233 A0A1S3KIH5 A0A1B6EM83 A0A1A7YW09 A0A2U9C1Q3 A0A195DN31 A0A1A8I8S2 A0A1A8AWY1 A0A1A8MD12 A0A3B4VF81 A0A182P5E7 A0A3B4VG43 A0A0J7KWM5 J3JX82 A0A2Y9D1B8 A0A3F2YZM4 A0A182V2F2 A0A182X5T2 Q7QHA7 A0A182HW63 A0A023EF22 A0A2U9C4A2 A0A1A8CL62 A0A1A8AEU7 N6STT7 A0A1A8HR78 A0A1B6GN29 A0A1Y1MPP9 A0A0S7M3S0 A0A3B4Y0V3 A0A182WDP7 E2A1C0 A0A1Y1MRM2 A0A1B6GUB1 I3JY28 A0A182YAX1 A0A3B4EQ68 A0A3P8R5Z7 A0A3P9CFQ2 A0A1L8DU31 A0A3P8UHR5 A0A1W4X8G7 A0A182R6M3 A0A084W8B9 A0A3B4Z9H0 A0A3P8R654 A0A3F2YVC4
S4PWY8 A0A194Q0N9 A0A212FH36 R4WCS3 A0A2R7VZZ6 A0A069DP08 A0A0N7Z8W8 R4FME4 A0A023F874 A0A0K8SAU0 A0A0A9XTG8 A0A0A9XNM0 A0A0K8S3W8 A0A0T6ATB1 A0A1B6GJ49 H2M9V6 A0A3Q0IJ42 A0A1S3CTP4 A0A3P9H2F3 Q1HQT9 A0A1S4E6Y6 A0A1B6LAX0 A0A3P9K4W8 J9HGS4 A0A3P8ZFE4 A0A1B6D7Z4 A0A1Q3FDY3 A0A182QAI6 E0VX45 A0A1B6DU45 G3NQZ4 A0A3B3C1Z1 Q16HL7 B0WPR4 G3NQZ9 A0A0K8TNB9 T1D5F9 Q4S6P0 C3KJX2 A0A182N254 A0A1S3KIX1 A0A1S3KIH6 V5GAP1 A0A2P8ZNQ0 H3D465 A0A060X233 A0A1S3KIH5 A0A1B6EM83 A0A1A7YW09 A0A2U9C1Q3 A0A195DN31 A0A1A8I8S2 A0A1A8AWY1 A0A1A8MD12 A0A3B4VF81 A0A182P5E7 A0A3B4VG43 A0A0J7KWM5 J3JX82 A0A2Y9D1B8 A0A3F2YZM4 A0A182V2F2 A0A182X5T2 Q7QHA7 A0A182HW63 A0A023EF22 A0A2U9C4A2 A0A1A8CL62 A0A1A8AEU7 N6STT7 A0A1A8HR78 A0A1B6GN29 A0A1Y1MPP9 A0A0S7M3S0 A0A3B4Y0V3 A0A182WDP7 E2A1C0 A0A1Y1MRM2 A0A1B6GUB1 I3JY28 A0A182YAX1 A0A3B4EQ68 A0A3P8R5Z7 A0A3P9CFQ2 A0A1L8DU31 A0A3P8UHR5 A0A1W4X8G7 A0A182R6M3 A0A084W8B9 A0A3B4Z9H0 A0A3P8R654 A0A3F2YVC4
PDB
3UIW
E-value=4.24944e-27,
Score=295
Ontologies
GO
Topology
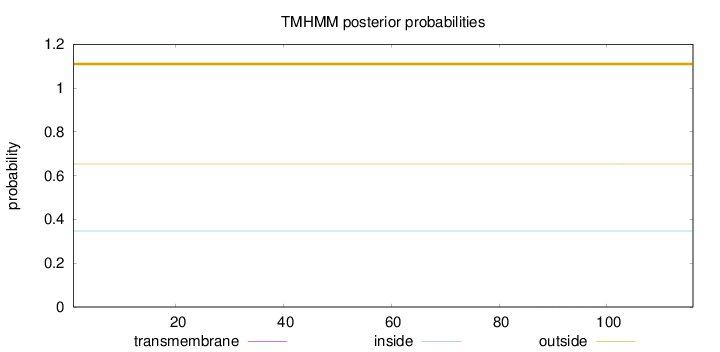
Length:
116
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00307
Exp number, first 60 AAs:
0
Total prob of N-in:
0.34708
outside
1 - 116
Population Genetic Test Statistics
Pi
352.249988
Theta
212.905805
Tajima's D
2.237644
CLR
0.079142
CSRT
0.92000399980001
Interpretation
Uncertain
