Pre Gene Modal
BGIBMGA009004
Annotation
PREDICTED:_uncharacterized_protein_LOC106142237_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.359
Sequence
CDS
ATGAATAGAAACTACAGTAGAAGAACCGCGGAACTCCACGATGAACACTGCTGCGCGGCCAATGTCAGCGGTAACTTTCGACGCAGCTCATCATTGCGGCTGCGCGGCGAAAAGATGGTACAGCGTTCACCACTTTGCACAAGGAAGTTGATTCCAATAATCACGGAAAACACGCCACAAAAACAACGCAGCGATGGTTCTCGACTCCAAGAGCCGGGTCATCGCCAGCGGAGTCATTCATTTAATTCAAATGCTCAGCAAAAGCCCAAAAAGTCCTGTTTAAAAACACAAGATGCTTGTATAGATCGAAACTTGAGCATGACTGATTCTGCCCACACACCACCAGGGTCACCTGAAGACTTACCTGATGATGAATCTCTGCATAGCTATGGGAGTGCAGCTACAGCTGCCTCTGTTGATGCTGGGTATGCTGCGTTCAATGGGACCACATTCAGTGGCAGATCAATGCGCTATGTTTTGCATTGCTCATCTCATGCAGGTTTGGCAGGTGATGAATATCTGACTCCTACACAGAGAGCTCAAAAGCAAATCCGTCGGCTTAAAAGTATGCTTAGTCAAGCAAAGAGAGACCTGGATAAAAAAGATAGTGAAATATTCCAACTAACAAAAGAAGTAGTTGAACTACGTCTTTATAAAACATCTATATGCTCACCTGACGAAAAATCAACCTCTAGTGAAATAGTTACTATAAGGGAGAATGCTGAAGATGCATTACATAATCAATCAACTTCTAAATGTGAAAAATCTTGTAGAAATTTTGATGTTATTGACAGCCCATTGTTTAAAGATGAAACACCAACAAGATGTCGCAATGAAATGCAAGGTTCGTTCACTGATTCTGGACATTTTGATGATCTCACAAACTCATCTTTACACTCTAAAGAATCTGTTCATATGCTTACACATGAAGTCTCTTGCATGACTGATATAAAGAATGCAGAGGAGGAAAGACATAGTTTGATTGCATTTTATGAGAAAAAAATTGAGGATGTTGTTCGCTCTCATGTTGGAGAAACACAAGAACTTAAAAAAGCTCATAATGACAAAGTAGAAGCCTTACTCCAGAAATTGGCTGATGTTAACACACGGTATTGTGAATTGCTACCTAATTATGAACAAGCAAAGGAAAGAATTCATTCCTTGGAAAAACAACTGGAAGAAGCAAGTAAGCAGTTAGAGGATGAAGATGGTAGAAACCGTTCAATGTACTTGCAGATGTACAACAAAGGTGTAGAAGCTGCTAAGTTTGAAATGGGGAAGGAGATTGAACCTGGAATGTCACAAGGGCAAATGAGCCGAGTTTCGGTGGAAGAGCTACTGGAGCAATTACAGATTACGCAGACTGAGCTCGAGAAAATCCGAGACACGGCATTCACTGAGGACCGAACGGCAAAATCACAAGTACTGCTAAGTGCAAAGGAGGCTGTTTCTTTATGGGTCCTAGGAGCTCGAAAGGCAATGTACCGTCGCATTGTCGAGGCGCAGAAAGGTAACAAAAACAACATCGATCCCGAAGTGACTCTCCAGTTTCTGAAATCTGCCATCTACTACTTCTTGACAGATCCTGAAAACCATCAAGGACATCTAAACGCTATAGAAAATATACTGGGATTTACTGAAGCCGAAAAGAAAAATATTCGCAAGGCAAGATCGACTTAG
Protein
MNRNYSRRTAELHDEHCCAANVSGNFRRSSSLRLRGEKMVQRSPLCTRKLIPIITENTPQKQRSDGSRLQEPGHRQRSHSFNSNAQQKPKKSCLKTQDACIDRNLSMTDSAHTPPGSPEDLPDDESLHSYGSAATAASVDAGYAAFNGTTFSGRSMRYVLHCSSHAGLAGDEYLTPTQRAQKQIRRLKSMLSQAKRDLDKKDSEIFQLTKEVVELRLYKTSICSPDEKSTSSEIVTIRENAEDALHNQSTSKCEKSCRNFDVIDSPLFKDETPTRCRNEMQGSFTDSGHFDDLTNSSLHSKESVHMLTHEVSCMTDIKNAEEERHSLIAFYEKKIEDVVRSHVGETQELKKAHNDKVEALLQKLADVNTRYCELLPNYEQAKERIHSLEKQLEEASKQLEDEDGRNRSMYLQMYNKGVEAAKFEMGKEIEPGMSQGQMSRVSVEELLEQLQITQTELEKIRDTAFTEDRTAKSQVLLSAKEAVSLWVLGARKAMYRRIVEAQKGNKNNIDPEVTLQFLKSAIYYFLTDPENHQGHLNAIENILGFTEAEKKNIRKARST
Summary
Uniprot
H9JHK6
A0A2H1V4Y1
A0A2A4JY80
A0A2A4JXM2
A0A194RMK1
A0A194PUH8
+ More
A0A212EQA2 A0A2A4JX42 A0A2A4JXG3 A0A2W1BV97 A0A0L7LB37 A0A1E1WIX4 A0A023ESQ1 A0A182Q9Y5 A0A182WFI4 A0A1S4G1Z1 A0A182MVI6 A0A182KA42 A0A1I8JU79 A0A182SW15 A0A2M4BKE3 W5J7L8 A0A2M4AH90 A0A2M4AHP9 Q7Q8I4 A0A182G7E3 A0A2Y9D0Q5 A0A1I8JU58 A0A2M3ZG20 A0A2M4BEV6 A0A2M3Z0P7 A0A2M4BF00 A0A2M4CTU1 A0A1S4GZH5 A0A084WFX7 A0A2M4AIU2 B0WLK4 A0A1Q3FZT5 A0A1Q3FZT3 A0A1J1ISP3 A0A1J1IW25 Q16G88 A0A2M3Z270 A0A2M4AHU0 A0A2M4AHH8 A0A182P4A8 W8BQ71 W8BJR5 A0A1B0CKJ9 A0A2J7PDE1 A0A182UZC7 A0A182X5Y4 A0A182IBL6 A0A034VEF4 A0A182NTD7 A0A182JJP2 A0A0A1X8K8 A0A182F6A8 A0A3B0JHM7 A0A182YN46 A0A2M4CSM9 A0A2M4CTX3 A0A2M4AD18 A0A0A1X520 A0A0P9A5U4 A0A1W4UIN0 A0A1W4UVX4 Q8I0H9 Q7KTP6 A0A0P8XGD2 A0A1B0BSC7 B4MTP9 A0A0J9QWZ0 M9PC20 A0A0R3NTX5 B4KGS9 A0A1Y1N7J9 A0A0J9QW01 B4I1C9 A0A0P8XG05 B4LUF3 Q8IPL0 A0A0J9QW19 A0A0J9QX00 A0A3B0KA50 A0A1Y1N808 A0A1W4UXB1 A0A0Q9WA92 B3MP99 A0A1W4UIM5 A0A1A9Z0D4 A0A0J9QW28 A0A0Q9WM51 A0A3B0K1Z8 A0A0K8TXG1 A0A0K8VN93 A0A0Q5W9F7 A0A1Q3FZX0 A0A1Q3FZV1 A0A0Q5WLL3
A0A212EQA2 A0A2A4JX42 A0A2A4JXG3 A0A2W1BV97 A0A0L7LB37 A0A1E1WIX4 A0A023ESQ1 A0A182Q9Y5 A0A182WFI4 A0A1S4G1Z1 A0A182MVI6 A0A182KA42 A0A1I8JU79 A0A182SW15 A0A2M4BKE3 W5J7L8 A0A2M4AH90 A0A2M4AHP9 Q7Q8I4 A0A182G7E3 A0A2Y9D0Q5 A0A1I8JU58 A0A2M3ZG20 A0A2M4BEV6 A0A2M3Z0P7 A0A2M4BF00 A0A2M4CTU1 A0A1S4GZH5 A0A084WFX7 A0A2M4AIU2 B0WLK4 A0A1Q3FZT5 A0A1Q3FZT3 A0A1J1ISP3 A0A1J1IW25 Q16G88 A0A2M3Z270 A0A2M4AHU0 A0A2M4AHH8 A0A182P4A8 W8BQ71 W8BJR5 A0A1B0CKJ9 A0A2J7PDE1 A0A182UZC7 A0A182X5Y4 A0A182IBL6 A0A034VEF4 A0A182NTD7 A0A182JJP2 A0A0A1X8K8 A0A182F6A8 A0A3B0JHM7 A0A182YN46 A0A2M4CSM9 A0A2M4CTX3 A0A2M4AD18 A0A0A1X520 A0A0P9A5U4 A0A1W4UIN0 A0A1W4UVX4 Q8I0H9 Q7KTP6 A0A0P8XGD2 A0A1B0BSC7 B4MTP9 A0A0J9QWZ0 M9PC20 A0A0R3NTX5 B4KGS9 A0A1Y1N7J9 A0A0J9QW01 B4I1C9 A0A0P8XG05 B4LUF3 Q8IPL0 A0A0J9QW19 A0A0J9QX00 A0A3B0KA50 A0A1Y1N808 A0A1W4UXB1 A0A0Q9WA92 B3MP99 A0A1W4UIM5 A0A1A9Z0D4 A0A0J9QW28 A0A0Q9WM51 A0A3B0K1Z8 A0A0K8TXG1 A0A0K8VN93 A0A0Q5W9F7 A0A1Q3FZX0 A0A1Q3FZV1 A0A0Q5WLL3
Pubmed
EMBL
BABH01012756
ODYU01000688
SOQ35887.1
NWSH01000444
PCG76380.1
PCG76384.1
+ More
KQ459989 KPJ18752.1 KQ459591 KPI96977.1 AGBW02013301 OWR43641.1 PCG76379.1 PCG76378.1 KZ149894 PZC78899.1 JTDY01001866 KOB72687.1 GDQN01004095 JAT86959.1 GAPW01001283 JAC12315.1 AXCN02000712 AXCM01002427 GGFJ01004389 MBW53530.1 ADMH02002020 ETN59961.1 GGFK01006657 MBW39978.1 GGFK01006992 MBW40313.1 AAAB01008944 EAA10063.3 JXUM01046476 JXUM01046477 JXUM01046478 KQ561478 KXJ78466.1 APCN01000708 GGFM01006680 MBW27431.1 GGFJ01002433 MBW51574.1 GGFM01001334 MBW22085.1 GGFJ01002432 MBW51573.1 GGFL01004070 MBW68248.1 ATLV01023407 KE525343 KFB49121.1 GGFK01007331 MBW40652.1 DS231987 EDS30534.1 GFDL01001978 JAV33067.1 GFDL01001980 JAV33065.1 CVRI01000058 CRL02746.1 CRL02745.1 CH478312 EAT33254.1 GGFM01001850 MBW22601.1 GGFK01006857 MBW40178.1 GGFK01006913 MBW40234.1 GAMC01007612 JAB98943.1 GAMC01007613 JAB98942.1 AJWK01016342 AJWK01016343 NEVH01026389 PNF14351.1 GAKP01019009 JAC39943.1 GBXI01006845 JAD07447.1 OUUW01000006 SPP81914.1 GGFL01004101 MBW68279.1 GGFL01004100 MBW68278.1 GGFK01005329 MBW38650.1 GBXI01008080 JAD06212.1 CH902620 KPU73847.1 KPU73849.1 AE014134 BT100049 AAN10527.1 ACX61590.1 AAN10529.2 KPU73848.1 JXJN01019668 JXJN01019669 JXJN01019670 CH963852 EDW75488.2 CM002910 KMY88239.1 AGB92619.1 CH379061 KRT04657.1 CH933807 EDW11129.2 GEZM01010756 JAV93851.1 KMY88237.1 CH480820 EDW54336.1 KPU73850.1 CH940649 EDW64139.2 AAN10528.2 KMY88236.1 KMY88243.1 KMY88249.1 SPP81911.1 GEZM01010758 JAV93849.1 KRF81439.1 KRF81442.1 EDV32218.2 KMY88246.1 KRF81440.1 SPP81910.1 GDHF01033579 GDHF01019218 GDHF01011971 JAI18735.1 JAI33096.1 JAI40343.1 GDHF01012001 JAI40313.1 CH954177 KQS70153.1 KQS70157.1 GFDL01001983 JAV33062.1 GFDL01001982 JAV33063.1 KQS70154.1
KQ459989 KPJ18752.1 KQ459591 KPI96977.1 AGBW02013301 OWR43641.1 PCG76379.1 PCG76378.1 KZ149894 PZC78899.1 JTDY01001866 KOB72687.1 GDQN01004095 JAT86959.1 GAPW01001283 JAC12315.1 AXCN02000712 AXCM01002427 GGFJ01004389 MBW53530.1 ADMH02002020 ETN59961.1 GGFK01006657 MBW39978.1 GGFK01006992 MBW40313.1 AAAB01008944 EAA10063.3 JXUM01046476 JXUM01046477 JXUM01046478 KQ561478 KXJ78466.1 APCN01000708 GGFM01006680 MBW27431.1 GGFJ01002433 MBW51574.1 GGFM01001334 MBW22085.1 GGFJ01002432 MBW51573.1 GGFL01004070 MBW68248.1 ATLV01023407 KE525343 KFB49121.1 GGFK01007331 MBW40652.1 DS231987 EDS30534.1 GFDL01001978 JAV33067.1 GFDL01001980 JAV33065.1 CVRI01000058 CRL02746.1 CRL02745.1 CH478312 EAT33254.1 GGFM01001850 MBW22601.1 GGFK01006857 MBW40178.1 GGFK01006913 MBW40234.1 GAMC01007612 JAB98943.1 GAMC01007613 JAB98942.1 AJWK01016342 AJWK01016343 NEVH01026389 PNF14351.1 GAKP01019009 JAC39943.1 GBXI01006845 JAD07447.1 OUUW01000006 SPP81914.1 GGFL01004101 MBW68279.1 GGFL01004100 MBW68278.1 GGFK01005329 MBW38650.1 GBXI01008080 JAD06212.1 CH902620 KPU73847.1 KPU73849.1 AE014134 BT100049 AAN10527.1 ACX61590.1 AAN10529.2 KPU73848.1 JXJN01019668 JXJN01019669 JXJN01019670 CH963852 EDW75488.2 CM002910 KMY88239.1 AGB92619.1 CH379061 KRT04657.1 CH933807 EDW11129.2 GEZM01010756 JAV93851.1 KMY88237.1 CH480820 EDW54336.1 KPU73850.1 CH940649 EDW64139.2 AAN10528.2 KMY88236.1 KMY88243.1 KMY88249.1 SPP81911.1 GEZM01010758 JAV93849.1 KRF81439.1 KRF81442.1 EDV32218.2 KMY88246.1 KRF81440.1 SPP81910.1 GDHF01033579 GDHF01019218 GDHF01011971 JAI18735.1 JAI33096.1 JAI40343.1 GDHF01012001 JAI40313.1 CH954177 KQS70153.1 KQS70157.1 GFDL01001983 JAV33062.1 GFDL01001982 JAV33063.1 KQS70154.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000075886 UP000075920 UP000075883 UP000075881 UP000075900 UP000075901 UP000000673 UP000007062 UP000069940 UP000249989 UP000075840 UP000030765 UP000002320 UP000183832 UP000008820 UP000075885 UP000092461 UP000235965 UP000075903 UP000076407 UP000075884 UP000075880 UP000069272 UP000268350 UP000076408 UP000007801 UP000192221 UP000000803 UP000092460 UP000007798 UP000001819 UP000009192 UP000001292 UP000008792 UP000092445 UP000008711
UP000075886 UP000075920 UP000075883 UP000075881 UP000075900 UP000075901 UP000000673 UP000007062 UP000069940 UP000249989 UP000075840 UP000030765 UP000002320 UP000183832 UP000008820 UP000075885 UP000092461 UP000235965 UP000075903 UP000076407 UP000075884 UP000075880 UP000069272 UP000268350 UP000076408 UP000007801 UP000192221 UP000000803 UP000092460 UP000007798 UP000001819 UP000009192 UP000001292 UP000008792 UP000092445 UP000008711
Pfam
PF01465 GRIP
SUPFAM
SSF140423
SSF140423
ProteinModelPortal
H9JHK6
A0A2H1V4Y1
A0A2A4JY80
A0A2A4JXM2
A0A194RMK1
A0A194PUH8
+ More
A0A212EQA2 A0A2A4JX42 A0A2A4JXG3 A0A2W1BV97 A0A0L7LB37 A0A1E1WIX4 A0A023ESQ1 A0A182Q9Y5 A0A182WFI4 A0A1S4G1Z1 A0A182MVI6 A0A182KA42 A0A1I8JU79 A0A182SW15 A0A2M4BKE3 W5J7L8 A0A2M4AH90 A0A2M4AHP9 Q7Q8I4 A0A182G7E3 A0A2Y9D0Q5 A0A1I8JU58 A0A2M3ZG20 A0A2M4BEV6 A0A2M3Z0P7 A0A2M4BF00 A0A2M4CTU1 A0A1S4GZH5 A0A084WFX7 A0A2M4AIU2 B0WLK4 A0A1Q3FZT5 A0A1Q3FZT3 A0A1J1ISP3 A0A1J1IW25 Q16G88 A0A2M3Z270 A0A2M4AHU0 A0A2M4AHH8 A0A182P4A8 W8BQ71 W8BJR5 A0A1B0CKJ9 A0A2J7PDE1 A0A182UZC7 A0A182X5Y4 A0A182IBL6 A0A034VEF4 A0A182NTD7 A0A182JJP2 A0A0A1X8K8 A0A182F6A8 A0A3B0JHM7 A0A182YN46 A0A2M4CSM9 A0A2M4CTX3 A0A2M4AD18 A0A0A1X520 A0A0P9A5U4 A0A1W4UIN0 A0A1W4UVX4 Q8I0H9 Q7KTP6 A0A0P8XGD2 A0A1B0BSC7 B4MTP9 A0A0J9QWZ0 M9PC20 A0A0R3NTX5 B4KGS9 A0A1Y1N7J9 A0A0J9QW01 B4I1C9 A0A0P8XG05 B4LUF3 Q8IPL0 A0A0J9QW19 A0A0J9QX00 A0A3B0KA50 A0A1Y1N808 A0A1W4UXB1 A0A0Q9WA92 B3MP99 A0A1W4UIM5 A0A1A9Z0D4 A0A0J9QW28 A0A0Q9WM51 A0A3B0K1Z8 A0A0K8TXG1 A0A0K8VN93 A0A0Q5W9F7 A0A1Q3FZX0 A0A1Q3FZV1 A0A0Q5WLL3
A0A212EQA2 A0A2A4JX42 A0A2A4JXG3 A0A2W1BV97 A0A0L7LB37 A0A1E1WIX4 A0A023ESQ1 A0A182Q9Y5 A0A182WFI4 A0A1S4G1Z1 A0A182MVI6 A0A182KA42 A0A1I8JU79 A0A182SW15 A0A2M4BKE3 W5J7L8 A0A2M4AH90 A0A2M4AHP9 Q7Q8I4 A0A182G7E3 A0A2Y9D0Q5 A0A1I8JU58 A0A2M3ZG20 A0A2M4BEV6 A0A2M3Z0P7 A0A2M4BF00 A0A2M4CTU1 A0A1S4GZH5 A0A084WFX7 A0A2M4AIU2 B0WLK4 A0A1Q3FZT5 A0A1Q3FZT3 A0A1J1ISP3 A0A1J1IW25 Q16G88 A0A2M3Z270 A0A2M4AHU0 A0A2M4AHH8 A0A182P4A8 W8BQ71 W8BJR5 A0A1B0CKJ9 A0A2J7PDE1 A0A182UZC7 A0A182X5Y4 A0A182IBL6 A0A034VEF4 A0A182NTD7 A0A182JJP2 A0A0A1X8K8 A0A182F6A8 A0A3B0JHM7 A0A182YN46 A0A2M4CSM9 A0A2M4CTX3 A0A2M4AD18 A0A0A1X520 A0A0P9A5U4 A0A1W4UIN0 A0A1W4UVX4 Q8I0H9 Q7KTP6 A0A0P8XGD2 A0A1B0BSC7 B4MTP9 A0A0J9QWZ0 M9PC20 A0A0R3NTX5 B4KGS9 A0A1Y1N7J9 A0A0J9QW01 B4I1C9 A0A0P8XG05 B4LUF3 Q8IPL0 A0A0J9QW19 A0A0J9QX00 A0A3B0KA50 A0A1Y1N808 A0A1W4UXB1 A0A0Q9WA92 B3MP99 A0A1W4UIM5 A0A1A9Z0D4 A0A0J9QW28 A0A0Q9WM51 A0A3B0K1Z8 A0A0K8TXG1 A0A0K8VN93 A0A0Q5W9F7 A0A1Q3FZX0 A0A1Q3FZV1 A0A0Q5WLL3
Ontologies
GO
PANTHER
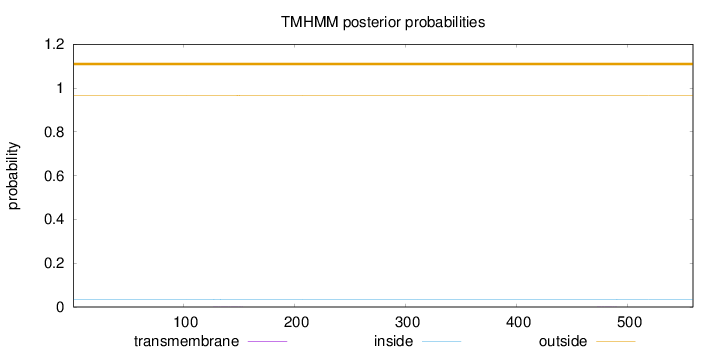
Topology
Length:
559
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02868
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03482
outside
1 - 559
Population Genetic Test Statistics
Pi
262.765918
Theta
183.145419
Tajima's D
1.256052
CLR
0.369551
CSRT
0.72336383180841
Interpretation
Uncertain
