Gene
KWMTBOMO01332
Pre Gene Modal
BGIBMGA008885
Annotation
PREDICTED:_protein_smoothened_isoform_X1_[Papilio_machaon]
Full name
Protein smoothened
Alternative Name
SMOH
Smooth
dSMO
Smooth
dSMO
Location in the cell
PlasmaMembrane Reliability : 4.914
Sequence
CDS
ATGATCTTGACGTGGACGCGGTGGCTGCTCGTCTTGTCATCATGTCGCGGGAGTCATTTCGATTCTGGCAGCGACAAAAACTTATTAGTGGACTTAAAGGAAAACGTCACCGTTCACCTTGAAGCGATTAACGGGACCCCAATTTACAGATTAATAAGACCAGACGAAAGTACACAATGGTTTCCGGAACGAGAAACGAAAATGAATAGTTGTGTGATGAGGGCCCGCTGTGAACCGCTTTTTAATTCCACGTGCCTCGGGTCGATACTTCCATATAGTAAAACAAGCGTGCACTTGACGTTCTACAATAGTCAGCGGCAAATACAGAACCAACTAGAGTTTTATAAAAATTTAATCAATGTACCCAATTGTTGGGCCGTTATCCAACCCTTGCTTTGTGCGACTTTCATGCCGAAATGTGAGAAAATACTAGATCACGATATGGTATATATGCCTTCGTTCGAGATGTGCAAAATAACAATGGAACCGTGCGCTATCCTCTATAACACTTCGTATTTTCCTTCTTTTCTCAAATGCAACACAACATTGTTTCCGCCGAATTGTGATAATGCCATGAGAGAGGTTAAATTTAATACAACCGGAAAATGTTTACCGCCCCTAATACACACGGACAAGTCCTTGCGCGTCTACGAAGGTATTACCGGTTGCGGGTTGCCGTGTCGTGACCCGTTGTATACTGAAGATGAACATCAACAAATACATAAACTGGTAGCCTGGGGCGCCGGTGTGTGTCTGGCCTTAAACCTGTTGACCGTAGCCACGTTCCTCATAGACTGGCGGAGTGCTAACAAATATCCCGCATTAGTTATATTTTATATCAACGTGTGTTTCGCTGTTGCATCAATGGGTTGGCTTGTTCAATTCGGTGTCGGCTCAAGAGACGATATAGTTTGCTCCAAAGATGGCACTAGACGACAAGGGGAGCCATCGGCCGAAGAAAATTTATTTTGCGTTGTCGTATTCGTTTTGGTTTATTATTTTATGATGGCAGCCTGCGTCTGGTTCGTTATATTCACGTACGCTTGGCACATGTGTAGTTTTCGAGCTTTGGGTAAAATACAAGATCGTATTGACAAGAAAGCTGCGTATTTCCACCTGGTGGCGTGGTCTCTCCCTTTGATATTAACGATAACAACAATGGCGTTCGGAGAAGTGGACGGGAGCAGCGTCACGGGCGTGTGCTTCGTCGGCTACGTGAACCATCCCATGCGCGCCGCCTTGTTGCTCGCGCCGCTGGCTGTCGTGCTTACTTTAGGCGGATATTTTCTTTTAAGAGGCGTATTTTCCCTTGTCGTGGTCCGAGTGTCGAGTAAAGACGTGATATCTCCGCGAGCCTCAAACAAGATCCGCCGTACGATCACGCGGTGCTCCATCACGGCGGCGCTGGTGGCCGTCTTCATCTGCGTCACGTTCGCTTGTCACATCTACGAGTTCAGGAATGCAGACCTTTGGAAAGACTCATTCAAGAGGAATATCATCTGTCGACTAGAAGCTGGTCAAGAAAGGCAGAAGGAATGTTTGACGGGCTCTCGTCCCTCGATATCTGTGCTGCAGCTGCGACTGCTGTGTTGCTTCGCTGCCGGTGCACTCATGTCCTCGTGGACCTGGACGCCGGCTGCCCTCGCCGCCTGGAGACGATATCTGGCCAGGAAATGTGGATGCTCCGTTGATGAGGAAATAACTCGTCGCGCACAAAAGCACGAGTTGATACGTCGAGCCTACAGACGCCGCGCGGAGTTCGCGGCCCGCGGACGGCTCTCCATATCTCTGGGCGGCTCCAGACAAGATCCCGTTGGGCTGTGTCTGGATCACACCTCTCCGATCGATTACCCGGAAGATGCGAAACATGAAAGCGGTGAATTGTCGTCGTCGTGGGCGGCCAACCTGCCGCGGTTCGTGCGGCGCCGCGACGCGCTCGTGTTGCCCGCGCACGCGCACCCGCACCACTCGCTCAGCTCCACGCCGGACCGACGCAACTCGCAGGACTCTCAGATCAGCATCAGCCTACGACACGTGTCCGTAGAATCTCGCCGCAACTCACTCGACAGTCAGCTCTCGGTGAAGATCGCCGAGATGAAAACCAAAGTCGGTCGCCGCCGAACCAAACATACCAAAGCGAAACGTAAACGGGCCACCGGGCCCCGCAAGGAGAGCACGCCTTCCATCGAGAGTCAAATAAGCCGCTATTGGCTGCAGGCCGTCGCGGCTAACAACGATCGCTCCAGAGAAGAAGTCAAATTCAGCTTTGAATGA
Protein
MILTWTRWLLVLSSCRGSHFDSGSDKNLLVDLKENVTVHLEAINGTPIYRLIRPDESTQWFPERETKMNSCVMRARCEPLFNSTCLGSILPYSKTSVHLTFYNSQRQIQNQLEFYKNLINVPNCWAVIQPLLCATFMPKCEKILDHDMVYMPSFEMCKITMEPCAILYNTSYFPSFLKCNTTLFPPNCDNAMREVKFNTTGKCLPPLIHTDKSLRVYEGITGCGLPCRDPLYTEDEHQQIHKLVAWGAGVCLALNLLTVATFLIDWRSANKYPALVIFYINVCFAVASMGWLVQFGVGSRDDIVCSKDGTRRQGEPSAEENLFCVVVFVLVYYFMMAACVWFVIFTYAWHMCSFRALGKIQDRIDKKAAYFHLVAWSLPLILTITTMAFGEVDGSSVTGVCFVGYVNHPMRAALLLAPLAVVLTLGGYFLLRGVFSLVVVRVSSKDVISPRASNKIRRTITRCSITAALVAVFICVTFACHIYEFRNADLWKDSFKRNIICRLEAGQERQKECLTGSRPSISVLQLRLLCCFAAGALMSSWTWTPAALAAWRRYLARKCGCSVDEEITRRAQKHELIRRAYRRRAEFAARGRLSISLGGSRQDPVGLCLDHTSPIDYPEDAKHESGELSSSWAANLPRFVRRRDALVLPAHAHPHHSLSSTPDRRNSQDSQISISLRHVSVESRRNSLDSQLSVKIAEMKTKVGRRRTKHTKAKRKRATGPRKESTPSIESQISRYWLQAVAANNDRSREEVKFSFE
Summary
Subunit
Interacts with cos.
Similarity
Belongs to the G-protein coupled receptor Fz/Smo family.
Keywords
3D-structure
Cell membrane
Complete proteome
Developmental protein
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Phosphoprotein
Receptor
Reference proteome
Signal
Transducer
Transmembrane
Transmembrane helix
Feature
chain Protein smoothened
Uniprot
A0A2W1BX54
A0A2A4JX22
A0A2A4JY61
A0A437AUY0
A0A212EQ67
A0A194RLP0
+ More
A0A194PVF9 A0A0L7LBB5 S4PKF0 A0A182YN57 Q175G5 A0A182RK65 A0A182TC12 A0A182MEB3 A0A182ILD3 A0A182GWI8 A0A182WFH4 A0A182HAH8 A0A182Q336 A0A084WFW9 A0A182P4A0 B0W046 A0A182F3D6 A0A182JRB6 A0A182MZ95 W5JQ90 A0A182VLW4 A0A1W4UQJ6 E5LCE7 A0A2Y9D159 A0A182X5X6 Q86PA9 D0QWD4 P91682 Q29NW6 B4GJP7 A0A182UA20 B4Q616 B4P2B8 B3N7N0 A0A0R1DIH5 B4ICU6 A0A0A1WQ87 B4MWJ5 A0A182KN72 A0A336K3M0 A0A3B0K8N5 A0A3B0JIG0 B4LU52 A0A1B0C9F1 B4JBG2 B3MUK2 A0A0K8UBW1 B4KHG7 A0A1I8PGR5 A0A1I8MLM7 A0A1S4GZI7 A0A0M4E766 Q7PPM7 A0A1A9WK74 A0A1I8MLM6 A0A1B0FJK7 A0A1B0AUL6 A0A1A9XQH4 A0A1A9UTD6 A0A1J1IML9 A0A2J7Q946 A0A2A3EJV5 A0A088APW7 A0A0L7R8D6 A0A154NZZ6 A0A0N0U309 T1HLP2 A0A224X7Q2 A0A023F248 A0A0K8TV70 E2B9T9 A0A158NQM4 A0A195D3P5 U5EP59 A0A195ERS4 A0A195BQZ3 A0A195DI48 F4WKG2 A0A151XB26 E2AN13 A0A0K8S5C7 A0A0A9ZEX8 A0A2R7WCF9 A0A034VSR5 A0A1Y1LLJ7 A0A1Y1LJ79 A0A232EKC5 K7IPU4 A0A026WDQ6 A0A067RTB7 A0A1B6DDX7 D6WXL7
A0A194PVF9 A0A0L7LBB5 S4PKF0 A0A182YN57 Q175G5 A0A182RK65 A0A182TC12 A0A182MEB3 A0A182ILD3 A0A182GWI8 A0A182WFH4 A0A182HAH8 A0A182Q336 A0A084WFW9 A0A182P4A0 B0W046 A0A182F3D6 A0A182JRB6 A0A182MZ95 W5JQ90 A0A182VLW4 A0A1W4UQJ6 E5LCE7 A0A2Y9D159 A0A182X5X6 Q86PA9 D0QWD4 P91682 Q29NW6 B4GJP7 A0A182UA20 B4Q616 B4P2B8 B3N7N0 A0A0R1DIH5 B4ICU6 A0A0A1WQ87 B4MWJ5 A0A182KN72 A0A336K3M0 A0A3B0K8N5 A0A3B0JIG0 B4LU52 A0A1B0C9F1 B4JBG2 B3MUK2 A0A0K8UBW1 B4KHG7 A0A1I8PGR5 A0A1I8MLM7 A0A1S4GZI7 A0A0M4E766 Q7PPM7 A0A1A9WK74 A0A1I8MLM6 A0A1B0FJK7 A0A1B0AUL6 A0A1A9XQH4 A0A1A9UTD6 A0A1J1IML9 A0A2J7Q946 A0A2A3EJV5 A0A088APW7 A0A0L7R8D6 A0A154NZZ6 A0A0N0U309 T1HLP2 A0A224X7Q2 A0A023F248 A0A0K8TV70 E2B9T9 A0A158NQM4 A0A195D3P5 U5EP59 A0A195ERS4 A0A195BQZ3 A0A195DI48 F4WKG2 A0A151XB26 E2AN13 A0A0K8S5C7 A0A0A9ZEX8 A0A2R7WCF9 A0A034VSR5 A0A1Y1LLJ7 A0A1Y1LJ79 A0A232EKC5 K7IPU4 A0A026WDQ6 A0A067RTB7 A0A1B6DDX7 D6WXL7
Pubmed
28756777
22118469
26354079
26227816
23622113
25244985
+ More
17510324 26483478 24438588 20920257 23761445 19859648 8706127 8700230 10731132 12537572 15616566 15691767 18327897 24351982 25639794 15632085 17994087 22936249 17550304 25830018 20966253 25315136 12364791 25474469 20798317 21347285 21719571 25401762 26823975 25348373 28004739 28648823 20075255 24508170 24845553 18362917 19820115
17510324 26483478 24438588 20920257 23761445 19859648 8706127 8700230 10731132 12537572 15616566 15691767 18327897 24351982 25639794 15632085 17994087 22936249 17550304 25830018 20966253 25315136 12364791 25474469 20798317 21347285 21719571 25401762 26823975 25348373 28004739 28648823 20075255 24508170 24845553 18362917 19820115
EMBL
KZ149894
PZC78901.1
NWSH01000444
PCG76359.1
PCG76360.1
RSAL01000400
+ More
RVE41928.1 AGBW02013301 OWR43640.1 KQ459989 KPJ18753.1 KQ459591 KPI96978.1 JTDY01001866 KOB72690.1 GAIX01004535 JAA88025.1 CH477401 EAT41704.2 AXCM01003295 JXUM01093223 JXUM01093224 KQ564042 KXJ72879.1 JXUM01031223 KQ560913 KXJ80372.1 AXCN02002069 ATLV01023407 KE525343 KFB49113.1 DS231816 EDS37618.1 ADMH02000838 ETN64904.1 HQ406725 ADQ00194.1 APCN01000708 BT003246 AAO25003.1 FJ821025 ACN94640.1 U87613 AF030334 AE014134 BT053691 CH379058 EAL34527.1 CH479184 EDW36863.1 CM000361 CM002910 EDX03200.1 KMY87280.1 CM000157 EDW87114.1 CH954177 EDV57206.1 KRJ97029.1 CH480829 EDW45372.1 GBXI01013270 JAD01022.1 CH963857 EDW76136.1 UFQS01000093 UFQT01000093 SSW99249.1 SSX19629.1 OUUW01000006 SPP81986.1 SPP81985.1 CH940649 EDW64039.2 AJWK01002277 AJWK01002278 CH916368 EDW03985.1 CH902624 EDV33531.2 GDHF01028253 JAI24061.1 CH933807 EDW11231.1 AAAB01008944 CP012523 ALC38552.1 EAA10160.2 CCAG010023359 JXJN01003674 CVRI01000055 CRL01467.1 NEVH01016943 PNF25083.1 KZ288232 PBC31542.1 KQ414632 KOC67135.1 KQ434782 KZC04568.1 KQ435922 KOX68621.1 ACPB03014790 GFTR01008050 JAW08376.1 GBBI01003708 JAC15004.1 GDHF01033940 JAI18374.1 GL446605 EFN87526.1 ADTU01023408 KQ976885 KYN07517.1 GANO01000299 JAB59572.1 KQ982021 KYN30579.1 KQ976419 KYM89276.1 KQ980824 KYN12516.1 GL888199 EGI65300.1 KQ982335 KYQ57539.1 GL441034 EFN65171.1 GBRD01017448 JAG48379.1 GBHO01001656 GDHC01005105 JAG41948.1 JAQ13524.1 KK854542 PTY16700.1 GAKP01014359 JAC44593.1 GEZM01054378 JAV73708.1 GEZM01054377 JAV73709.1 NNAY01003824 OXU18795.1 KK107260 EZA54187.1 KK852428 KDR24055.1 GEDC01013404 JAS23894.1 KQ971362 EFA09161.1
RVE41928.1 AGBW02013301 OWR43640.1 KQ459989 KPJ18753.1 KQ459591 KPI96978.1 JTDY01001866 KOB72690.1 GAIX01004535 JAA88025.1 CH477401 EAT41704.2 AXCM01003295 JXUM01093223 JXUM01093224 KQ564042 KXJ72879.1 JXUM01031223 KQ560913 KXJ80372.1 AXCN02002069 ATLV01023407 KE525343 KFB49113.1 DS231816 EDS37618.1 ADMH02000838 ETN64904.1 HQ406725 ADQ00194.1 APCN01000708 BT003246 AAO25003.1 FJ821025 ACN94640.1 U87613 AF030334 AE014134 BT053691 CH379058 EAL34527.1 CH479184 EDW36863.1 CM000361 CM002910 EDX03200.1 KMY87280.1 CM000157 EDW87114.1 CH954177 EDV57206.1 KRJ97029.1 CH480829 EDW45372.1 GBXI01013270 JAD01022.1 CH963857 EDW76136.1 UFQS01000093 UFQT01000093 SSW99249.1 SSX19629.1 OUUW01000006 SPP81986.1 SPP81985.1 CH940649 EDW64039.2 AJWK01002277 AJWK01002278 CH916368 EDW03985.1 CH902624 EDV33531.2 GDHF01028253 JAI24061.1 CH933807 EDW11231.1 AAAB01008944 CP012523 ALC38552.1 EAA10160.2 CCAG010023359 JXJN01003674 CVRI01000055 CRL01467.1 NEVH01016943 PNF25083.1 KZ288232 PBC31542.1 KQ414632 KOC67135.1 KQ434782 KZC04568.1 KQ435922 KOX68621.1 ACPB03014790 GFTR01008050 JAW08376.1 GBBI01003708 JAC15004.1 GDHF01033940 JAI18374.1 GL446605 EFN87526.1 ADTU01023408 KQ976885 KYN07517.1 GANO01000299 JAB59572.1 KQ982021 KYN30579.1 KQ976419 KYM89276.1 KQ980824 KYN12516.1 GL888199 EGI65300.1 KQ982335 KYQ57539.1 GL441034 EFN65171.1 GBRD01017448 JAG48379.1 GBHO01001656 GDHC01005105 JAG41948.1 JAQ13524.1 KK854542 PTY16700.1 GAKP01014359 JAC44593.1 GEZM01054378 JAV73708.1 GEZM01054377 JAV73709.1 NNAY01003824 OXU18795.1 KK107260 EZA54187.1 KK852428 KDR24055.1 GEDC01013404 JAS23894.1 KQ971362 EFA09161.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000076408 UP000008820 UP000075900 UP000075901 UP000075883 UP000075880 UP000069940 UP000249989 UP000075920 UP000075886 UP000030765 UP000075885 UP000002320 UP000069272 UP000075881 UP000075884 UP000000673 UP000075903 UP000192221 UP000075840 UP000076407 UP000000803 UP000001819 UP000008744 UP000075902 UP000000304 UP000002282 UP000008711 UP000001292 UP000007798 UP000075882 UP000268350 UP000008792 UP000092461 UP000001070 UP000007801 UP000009192 UP000095300 UP000095301 UP000092553 UP000007062 UP000091820 UP000092444 UP000092460 UP000092443 UP000078200 UP000183832 UP000235965 UP000242457 UP000005203 UP000053825 UP000076502 UP000053105 UP000015103 UP000008237 UP000005205 UP000078542 UP000078541 UP000078540 UP000078492 UP000007755 UP000075809 UP000000311 UP000215335 UP000002358 UP000053097 UP000027135 UP000007266
UP000076408 UP000008820 UP000075900 UP000075901 UP000075883 UP000075880 UP000069940 UP000249989 UP000075920 UP000075886 UP000030765 UP000075885 UP000002320 UP000069272 UP000075881 UP000075884 UP000000673 UP000075903 UP000192221 UP000075840 UP000076407 UP000000803 UP000001819 UP000008744 UP000075902 UP000000304 UP000002282 UP000008711 UP000001292 UP000007798 UP000075882 UP000268350 UP000008792 UP000092461 UP000001070 UP000007801 UP000009192 UP000095300 UP000095301 UP000092553 UP000007062 UP000091820 UP000092444 UP000092460 UP000092443 UP000078200 UP000183832 UP000235965 UP000242457 UP000005203 UP000053825 UP000076502 UP000053105 UP000015103 UP000008237 UP000005205 UP000078542 UP000078541 UP000078540 UP000078492 UP000007755 UP000075809 UP000000311 UP000215335 UP000002358 UP000053097 UP000027135 UP000007266
Interpro
IPR017981
GPCR_2-like
+ More
IPR036790 Frizzled_dom_sf
IPR000539 Frizzled/Smoothened_TM
IPR035683 SMO_7TM
IPR026544 SMO
IPR020067 Frizzled_dom
IPR015526 Frizzled/SFRP
IPR036322 WD40_repeat_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR041771 SMO_CRD
IPR022776 TRM13/UPF0224_CHHC_Znf_dom
IPR036236 Znf_C2H2_sf
IPR036790 Frizzled_dom_sf
IPR000539 Frizzled/Smoothened_TM
IPR035683 SMO_7TM
IPR026544 SMO
IPR020067 Frizzled_dom
IPR015526 Frizzled/SFRP
IPR036322 WD40_repeat_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR041771 SMO_CRD
IPR022776 TRM13/UPF0224_CHHC_Znf_dom
IPR036236 Znf_C2H2_sf
Gene 3D
ProteinModelPortal
A0A2W1BX54
A0A2A4JX22
A0A2A4JY61
A0A437AUY0
A0A212EQ67
A0A194RLP0
+ More
A0A194PVF9 A0A0L7LBB5 S4PKF0 A0A182YN57 Q175G5 A0A182RK65 A0A182TC12 A0A182MEB3 A0A182ILD3 A0A182GWI8 A0A182WFH4 A0A182HAH8 A0A182Q336 A0A084WFW9 A0A182P4A0 B0W046 A0A182F3D6 A0A182JRB6 A0A182MZ95 W5JQ90 A0A182VLW4 A0A1W4UQJ6 E5LCE7 A0A2Y9D159 A0A182X5X6 Q86PA9 D0QWD4 P91682 Q29NW6 B4GJP7 A0A182UA20 B4Q616 B4P2B8 B3N7N0 A0A0R1DIH5 B4ICU6 A0A0A1WQ87 B4MWJ5 A0A182KN72 A0A336K3M0 A0A3B0K8N5 A0A3B0JIG0 B4LU52 A0A1B0C9F1 B4JBG2 B3MUK2 A0A0K8UBW1 B4KHG7 A0A1I8PGR5 A0A1I8MLM7 A0A1S4GZI7 A0A0M4E766 Q7PPM7 A0A1A9WK74 A0A1I8MLM6 A0A1B0FJK7 A0A1B0AUL6 A0A1A9XQH4 A0A1A9UTD6 A0A1J1IML9 A0A2J7Q946 A0A2A3EJV5 A0A088APW7 A0A0L7R8D6 A0A154NZZ6 A0A0N0U309 T1HLP2 A0A224X7Q2 A0A023F248 A0A0K8TV70 E2B9T9 A0A158NQM4 A0A195D3P5 U5EP59 A0A195ERS4 A0A195BQZ3 A0A195DI48 F4WKG2 A0A151XB26 E2AN13 A0A0K8S5C7 A0A0A9ZEX8 A0A2R7WCF9 A0A034VSR5 A0A1Y1LLJ7 A0A1Y1LJ79 A0A232EKC5 K7IPU4 A0A026WDQ6 A0A067RTB7 A0A1B6DDX7 D6WXL7
A0A194PVF9 A0A0L7LBB5 S4PKF0 A0A182YN57 Q175G5 A0A182RK65 A0A182TC12 A0A182MEB3 A0A182ILD3 A0A182GWI8 A0A182WFH4 A0A182HAH8 A0A182Q336 A0A084WFW9 A0A182P4A0 B0W046 A0A182F3D6 A0A182JRB6 A0A182MZ95 W5JQ90 A0A182VLW4 A0A1W4UQJ6 E5LCE7 A0A2Y9D159 A0A182X5X6 Q86PA9 D0QWD4 P91682 Q29NW6 B4GJP7 A0A182UA20 B4Q616 B4P2B8 B3N7N0 A0A0R1DIH5 B4ICU6 A0A0A1WQ87 B4MWJ5 A0A182KN72 A0A336K3M0 A0A3B0K8N5 A0A3B0JIG0 B4LU52 A0A1B0C9F1 B4JBG2 B3MUK2 A0A0K8UBW1 B4KHG7 A0A1I8PGR5 A0A1I8MLM7 A0A1S4GZI7 A0A0M4E766 Q7PPM7 A0A1A9WK74 A0A1I8MLM6 A0A1B0FJK7 A0A1B0AUL6 A0A1A9XQH4 A0A1A9UTD6 A0A1J1IML9 A0A2J7Q946 A0A2A3EJV5 A0A088APW7 A0A0L7R8D6 A0A154NZZ6 A0A0N0U309 T1HLP2 A0A224X7Q2 A0A023F248 A0A0K8TV70 E2B9T9 A0A158NQM4 A0A195D3P5 U5EP59 A0A195ERS4 A0A195BQZ3 A0A195DI48 F4WKG2 A0A151XB26 E2AN13 A0A0K8S5C7 A0A0A9ZEX8 A0A2R7WCF9 A0A034VSR5 A0A1Y1LLJ7 A0A1Y1LJ79 A0A232EKC5 K7IPU4 A0A026WDQ6 A0A067RTB7 A0A1B6DDX7 D6WXL7
PDB
5V56
E-value=1.40453e-99,
Score=930
Ontologies
KEGG
GO
GO:0016021
GO:0004888
GO:0007275
GO:0007224
GO:0007166
GO:0070273
GO:0035019
GO:0007193
GO:0007346
GO:0048592
GO:0005929
GO:0048099
GO:1904263
GO:0043025
GO:0042803
GO:0001746
GO:2000134
GO:0030707
GO:0005887
GO:0007455
GO:0042802
GO:0042981
GO:0002385
GO:0007350
GO:0007476
GO:0007367
GO:0030425
GO:0004930
GO:1903078
GO:0048100
GO:0005886
GO:0030424
GO:0007417
GO:0071679
GO:0005113
GO:0045880
GO:0005515
GO:0016020
GO:0016820
GO:0016042
GO:0005634
GO:0006334
GO:0003779
PANTHER
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 17,
Likelihood: 0.794792
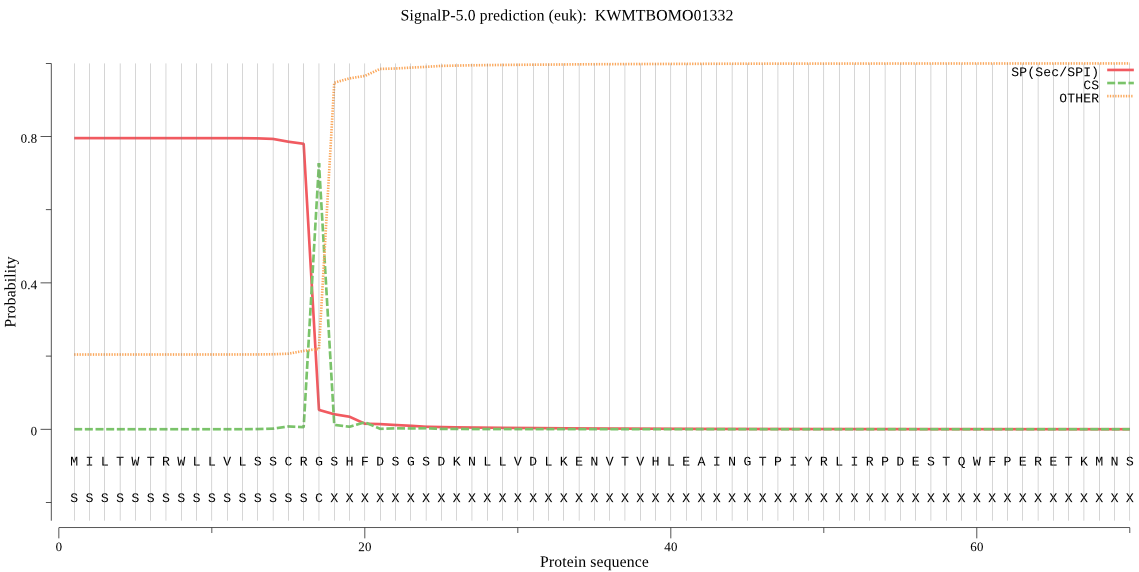
Length:
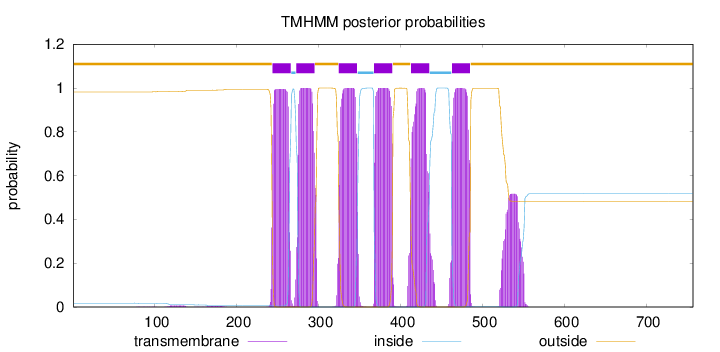
757
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
145.33112
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.01753
outside
1 - 243
TMhelix
244 - 266
inside
267 - 272
TMhelix
273 - 295
outside
296 - 324
TMhelix
325 - 347
inside
348 - 367
TMhelix
368 - 390
outside
391 - 412
TMhelix
413 - 435
inside
436 - 462
TMhelix
463 - 485
outside
486 - 757
Population Genetic Test Statistics
Pi
237.07767
Theta
194.183068
Tajima's D
0.834964
CLR
12.112843
CSRT
0.609519524023799
Interpretation
Uncertain
