Gene
KWMTBOMO01327
Pre Gene Modal
BGIBMGA009007
Annotation
PREDICTED:_dyslexia-associated_protein_KIAA0319_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.88
Sequence
CDS
ATGCTCACAAATATAGAAGCGGGGCGCTACGTGTTTGAACTGACGGTGACTGACGACCAAGGCGAGTCGGACAGGGACACTGTCAGTGTTCAAGTCAAACCGGACCCGCTCGAGATGAGCCTGCTTGAGATGACTCTGAATATCCCGGTATCTACATTTACGCAAGGACAGTTGGACTCGTTGGTACAAAAAATGGCTCTGCTCTTAAAAGGAGATGTCAAGATGAACGTAACAAAAGTATTCGGACAAGTCGACTCCGGGAATACACAGCTGATATTCTTTCTAACTGAAGACAATAAGGTGATGGAGGGTATACGCGCGTTGACGCTGTCCCGGGGCGCGGCTCGTGCAGGCGCGTTCGGAGGCGCGGCGCGGGTGCGTGCGCTGCGCTGTCTGAGCACGTGCTCGGGGCGCGGCCGCTGCGACCGCACCGACCACACGTGCCGCTGCGACGCCTTCTGGATGCAGAGCCTCTTCACGCAGCTCGACCCCGACGCCATGCCTAATTGCGTTCTAGAAAACCGTCCTGGATTTAACTAA
Protein
MLTNIEAGRYVFELTVTDDQGESDRDTVSVQVKPDPLEMSLLEMTLNIPVSTFTQGQLDSLVQKMALLLKGDVKMNVTKVFGQVDSGNTQLIFFLTEDNKVMEGIRALTLSRGAARAGAFGGAARVRALRCLSTCSGRGRCDRTDHTCRCDAFWMQSLFTQLDPDAMPNCVLENRPGFN
Summary
Uniprot
H9JHK9
A0A2W1BZN6
A0A2H1V4X0
A0A0N1PEV5
A0A194PUI2
A0A194RRS2
+ More
A0A212EQ76 A0A0L7LB05 A0A1B6CDV3 A0A1B6F1Z1 A0A1B6MA64 A0A1B6KMD1 A0A1B6DYW4 A0A0A9XSD2 A0A0A9XUH3 A0A0A9XZ38 A0A0K8TBS6 E2C738 A0A146LHA4 A0A0A9XZ45 A0A0A9XPA1 A0A0A9XSC8 A0A2S2QNI3 B4MXD2 A0A224XHV3 J9K213 A0A2A3EKI6 A0A069DZN1 A0A0P8YHW4 A0A2H8TF87 B3M431 A0A088APY5 A0A0C9R6L1 A0A195FRF2 A0A026VVX5 F4WC84 T1HMH1 A0A158N9Y4 B4IQA7 A0A195BK20 A0A0N0BCF2 A0A2S2PQQ2 A0A0L7R8G6 A0A195C3F4 B4KUX2 A0A067QRR4 A0A195ELE1 A0A151WHB5 A0A1W4VAD1 E0VU62 B4H8R1 Q29DN7 B4HIY7 A0A182GKZ5 A0A1Q3FF91 U5EVV3 B4QL00 B4J0Z7 A0A0J9RSJ4 Q16M97 A0A2P8YZ74 A0A3B0JYR9 A0A1Q3FBJ8 A0A1Q3FBJ4 A0A1Q3FAK2 A0A1Q3FAP7 B0W015 A0A3B0K1J8 A0A182GL46 Q9VSC9 F7E448 A0A0J7LAD4 A0A0M4ENB7 A0A336KZH6 A0A336N0I4 B4PCS1 B4LBZ7 K7IQ31 A0A232EQ44 T1GY78 D6WYN3 V5GRA1 A0A0L0C9U7 B3NF63 A0A2M4BCF2 A0A2M4BBD9 A0A2M4BBA1 A0A2M4BD80 A0A2M4ABB6 A0A2M4ABW3 W5JCZ2 A0A0T6B207 A0A182F361 A0A2M3Z5B1 A0A1B0D666 A0A1I8NB24 A0A182WFF8 Q7Q8G8
A0A212EQ76 A0A0L7LB05 A0A1B6CDV3 A0A1B6F1Z1 A0A1B6MA64 A0A1B6KMD1 A0A1B6DYW4 A0A0A9XSD2 A0A0A9XUH3 A0A0A9XZ38 A0A0K8TBS6 E2C738 A0A146LHA4 A0A0A9XZ45 A0A0A9XPA1 A0A0A9XSC8 A0A2S2QNI3 B4MXD2 A0A224XHV3 J9K213 A0A2A3EKI6 A0A069DZN1 A0A0P8YHW4 A0A2H8TF87 B3M431 A0A088APY5 A0A0C9R6L1 A0A195FRF2 A0A026VVX5 F4WC84 T1HMH1 A0A158N9Y4 B4IQA7 A0A195BK20 A0A0N0BCF2 A0A2S2PQQ2 A0A0L7R8G6 A0A195C3F4 B4KUX2 A0A067QRR4 A0A195ELE1 A0A151WHB5 A0A1W4VAD1 E0VU62 B4H8R1 Q29DN7 B4HIY7 A0A182GKZ5 A0A1Q3FF91 U5EVV3 B4QL00 B4J0Z7 A0A0J9RSJ4 Q16M97 A0A2P8YZ74 A0A3B0JYR9 A0A1Q3FBJ8 A0A1Q3FBJ4 A0A1Q3FAK2 A0A1Q3FAP7 B0W015 A0A3B0K1J8 A0A182GL46 Q9VSC9 F7E448 A0A0J7LAD4 A0A0M4ENB7 A0A336KZH6 A0A336N0I4 B4PCS1 B4LBZ7 K7IQ31 A0A232EQ44 T1GY78 D6WYN3 V5GRA1 A0A0L0C9U7 B3NF63 A0A2M4BCF2 A0A2M4BBD9 A0A2M4BBA1 A0A2M4BD80 A0A2M4ABB6 A0A2M4ABW3 W5JCZ2 A0A0T6B207 A0A182F361 A0A2M3Z5B1 A0A1B0D666 A0A1I8NB24 A0A182WFF8 Q7Q8G8
Pubmed
19121390
28756777
26354079
22118469
26227816
25401762
+ More
20798317 26823975 17994087 26334808 24508170 30249741 21719571 21347285 24845553 20566863 15632085 26483478 22936249 17510324 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20431018 17550304 20075255 28648823 18362917 19820115 26108605 20920257 23761445 25315136 12364791
20798317 26823975 17994087 26334808 24508170 30249741 21719571 21347285 24845553 20566863 15632085 26483478 22936249 17510324 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20431018 17550304 20075255 28648823 18362917 19820115 26108605 20920257 23761445 25315136 12364791
EMBL
BABH01012760
BABH01012761
BABH01012762
KZ149894
PZC78905.1
ODYU01000688
+ More
SOQ35883.1 KQ459273 KPJ02053.1 KQ459591 KPI96982.1 KQ459989 KPJ18756.1 AGBW02013301 OWR43636.1 JTDY01001898 KOB72590.1 GEDC01025664 JAS11634.1 GECZ01025602 JAS44167.1 GEBQ01007155 JAT32822.1 GEBQ01027623 JAT12354.1 GEDC01006440 GEDC01004669 JAS30858.1 JAS32629.1 GBHO01021043 JAG22561.1 GBHO01021046 JAG22558.1 GBHO01021049 JAG22555.1 GBRD01003211 JAG62610.1 GL453303 EFN76237.1 GDHC01011171 JAQ07458.1 GBHO01021044 JAG22560.1 GBHO01021047 JAG22557.1 GBHO01021048 JAG22556.1 GGMS01009907 MBY79110.1 CH963876 EDW76701.2 GFTR01008396 JAW08030.1 ABLF02027275 ABLF02027279 KZ288232 PBC31541.1 GBGD01000350 JAC88539.1 CH902618 KPU78543.1 GFXV01000958 MBW12763.1 EDV40393.1 GBYB01011969 JAG81736.1 KQ981305 KYN42862.1 KK107796 QOIP01000013 EZA47636.1 RLU15463.1 GL888070 EGI68064.1 ACPB03008516 ADTU01009910 CH686588 EDW44130.1 KQ976455 KYM85042.1 KQ435922 KOX68622.1 GGMR01019116 MBY31735.1 KQ414632 KOC67134.1 KQ978292 KYM95382.1 CH933809 EDW18283.2 KK853016 KDR12486.1 KQ978739 KYN28752.1 KQ983135 KYQ47195.1 DS235780 EEB16918.1 CH479224 EDW35104.1 CH379070 EAL30377.2 CH480815 EDW40641.1 JXUM01070969 JXUM01070970 JXUM01070971 JXUM01070972 JXUM01070973 JXUM01070974 KQ562637 KXJ75444.1 GFDL01008825 JAV26220.1 GANO01001702 JAB58169.1 CM000363 EDX09633.1 CH916366 EDV96852.1 CM002912 KMY98274.1 CH477870 EAT35458.1 PYGN01000274 PSN49545.1 OUUW01000012 SPP87194.1 GFDL01010156 JAV24889.1 GFDL01010115 JAV24930.1 GFDL01010430 JAV24615.1 GFDL01010428 JAV24617.1 DS231816 EDS37587.1 SPP87193.1 JXUM01071235 JXUM01071236 JXUM01071237 KQ562651 KXJ75399.1 AE014296 BT133232 AAF50494.1 AFA36415.1 AAMC01058419 AAMC01058420 LBMM01000051 KMR05151.1 CP012525 ALC43405.1 UFQS01001424 UFQT01001424 SSX10814.1 SSX30495.1 UFQT01002870 SSX34197.1 CM000159 EDW93825.1 CH940647 EDW69797.2 NNAY01002839 OXU20461.1 CAQQ02382422 KQ971357 EFA08418.1 GALX01004339 JAB64127.1 JRES01000714 KNC29026.1 CH954178 EDV50405.1 GGFJ01001584 MBW50725.1 GGFJ01001160 MBW50301.1 GGFJ01001159 MBW50300.1 GGFJ01001637 MBW50778.1 GGFK01004762 MBW38083.1 GGFK01004777 MBW38098.1 ADMH02001597 ETN61916.1 LJIG01016176 KRT81386.1 GGFM01002956 MBW23707.1 AJVK01011992 AAAB01008944 EAA10200.4
SOQ35883.1 KQ459273 KPJ02053.1 KQ459591 KPI96982.1 KQ459989 KPJ18756.1 AGBW02013301 OWR43636.1 JTDY01001898 KOB72590.1 GEDC01025664 JAS11634.1 GECZ01025602 JAS44167.1 GEBQ01007155 JAT32822.1 GEBQ01027623 JAT12354.1 GEDC01006440 GEDC01004669 JAS30858.1 JAS32629.1 GBHO01021043 JAG22561.1 GBHO01021046 JAG22558.1 GBHO01021049 JAG22555.1 GBRD01003211 JAG62610.1 GL453303 EFN76237.1 GDHC01011171 JAQ07458.1 GBHO01021044 JAG22560.1 GBHO01021047 JAG22557.1 GBHO01021048 JAG22556.1 GGMS01009907 MBY79110.1 CH963876 EDW76701.2 GFTR01008396 JAW08030.1 ABLF02027275 ABLF02027279 KZ288232 PBC31541.1 GBGD01000350 JAC88539.1 CH902618 KPU78543.1 GFXV01000958 MBW12763.1 EDV40393.1 GBYB01011969 JAG81736.1 KQ981305 KYN42862.1 KK107796 QOIP01000013 EZA47636.1 RLU15463.1 GL888070 EGI68064.1 ACPB03008516 ADTU01009910 CH686588 EDW44130.1 KQ976455 KYM85042.1 KQ435922 KOX68622.1 GGMR01019116 MBY31735.1 KQ414632 KOC67134.1 KQ978292 KYM95382.1 CH933809 EDW18283.2 KK853016 KDR12486.1 KQ978739 KYN28752.1 KQ983135 KYQ47195.1 DS235780 EEB16918.1 CH479224 EDW35104.1 CH379070 EAL30377.2 CH480815 EDW40641.1 JXUM01070969 JXUM01070970 JXUM01070971 JXUM01070972 JXUM01070973 JXUM01070974 KQ562637 KXJ75444.1 GFDL01008825 JAV26220.1 GANO01001702 JAB58169.1 CM000363 EDX09633.1 CH916366 EDV96852.1 CM002912 KMY98274.1 CH477870 EAT35458.1 PYGN01000274 PSN49545.1 OUUW01000012 SPP87194.1 GFDL01010156 JAV24889.1 GFDL01010115 JAV24930.1 GFDL01010430 JAV24615.1 GFDL01010428 JAV24617.1 DS231816 EDS37587.1 SPP87193.1 JXUM01071235 JXUM01071236 JXUM01071237 KQ562651 KXJ75399.1 AE014296 BT133232 AAF50494.1 AFA36415.1 AAMC01058419 AAMC01058420 LBMM01000051 KMR05151.1 CP012525 ALC43405.1 UFQS01001424 UFQT01001424 SSX10814.1 SSX30495.1 UFQT01002870 SSX34197.1 CM000159 EDW93825.1 CH940647 EDW69797.2 NNAY01002839 OXU20461.1 CAQQ02382422 KQ971357 EFA08418.1 GALX01004339 JAB64127.1 JRES01000714 KNC29026.1 CH954178 EDV50405.1 GGFJ01001584 MBW50725.1 GGFJ01001160 MBW50301.1 GGFJ01001159 MBW50300.1 GGFJ01001637 MBW50778.1 GGFK01004762 MBW38083.1 GGFK01004777 MBW38098.1 ADMH02001597 ETN61916.1 LJIG01016176 KRT81386.1 GGFM01002956 MBW23707.1 AJVK01011992 AAAB01008944 EAA10200.4
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000008237
+ More
UP000007798 UP000007819 UP000242457 UP000007801 UP000005203 UP000078541 UP000053097 UP000279307 UP000007755 UP000015103 UP000005205 UP000001292 UP000078540 UP000053105 UP000053825 UP000078542 UP000009192 UP000027135 UP000078492 UP000075809 UP000192221 UP000009046 UP000008744 UP000001819 UP000069940 UP000249989 UP000000304 UP000001070 UP000008820 UP000245037 UP000268350 UP000002320 UP000000803 UP000008143 UP000036403 UP000092553 UP000002282 UP000008792 UP000002358 UP000215335 UP000015102 UP000007266 UP000037069 UP000008711 UP000000673 UP000069272 UP000092462 UP000095301 UP000075920 UP000007062
UP000007798 UP000007819 UP000242457 UP000007801 UP000005203 UP000078541 UP000053097 UP000279307 UP000007755 UP000015103 UP000005205 UP000001292 UP000078540 UP000053105 UP000053825 UP000078542 UP000009192 UP000027135 UP000078492 UP000075809 UP000192221 UP000009046 UP000008744 UP000001819 UP000069940 UP000249989 UP000000304 UP000001070 UP000008820 UP000245037 UP000268350 UP000002320 UP000000803 UP000008143 UP000036403 UP000092553 UP000002282 UP000008792 UP000002358 UP000215335 UP000015102 UP000007266 UP000037069 UP000008711 UP000000673 UP000069272 UP000092462 UP000095301 UP000075920 UP000007062
Interpro
IPR011106
MANSC_N
+ More
IPR022409 PKD/Chitinase_dom
IPR013783 Ig-like_fold
IPR035986 PKD_dom_sf
IPR013980 MANSC_dom
IPR003599 Ig_sub
IPR000601 PKD_dom
IPR013032 EGF-like_CS
IPR000315 Znf_B-box
IPR009030 Growth_fac_rcpt_cys_sf
IPR000742 EGF-like_dom
IPR001841 Znf_RING
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR002859 PKD/REJ-like
IPR003961 FN3_dom
IPR004279 Perilipin
IPR022409 PKD/Chitinase_dom
IPR013783 Ig-like_fold
IPR035986 PKD_dom_sf
IPR013980 MANSC_dom
IPR003599 Ig_sub
IPR000601 PKD_dom
IPR013032 EGF-like_CS
IPR000315 Znf_B-box
IPR009030 Growth_fac_rcpt_cys_sf
IPR000742 EGF-like_dom
IPR001841 Znf_RING
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR002859 PKD/REJ-like
IPR003961 FN3_dom
IPR004279 Perilipin
Gene 3D
ProteinModelPortal
H9JHK9
A0A2W1BZN6
A0A2H1V4X0
A0A0N1PEV5
A0A194PUI2
A0A194RRS2
+ More
A0A212EQ76 A0A0L7LB05 A0A1B6CDV3 A0A1B6F1Z1 A0A1B6MA64 A0A1B6KMD1 A0A1B6DYW4 A0A0A9XSD2 A0A0A9XUH3 A0A0A9XZ38 A0A0K8TBS6 E2C738 A0A146LHA4 A0A0A9XZ45 A0A0A9XPA1 A0A0A9XSC8 A0A2S2QNI3 B4MXD2 A0A224XHV3 J9K213 A0A2A3EKI6 A0A069DZN1 A0A0P8YHW4 A0A2H8TF87 B3M431 A0A088APY5 A0A0C9R6L1 A0A195FRF2 A0A026VVX5 F4WC84 T1HMH1 A0A158N9Y4 B4IQA7 A0A195BK20 A0A0N0BCF2 A0A2S2PQQ2 A0A0L7R8G6 A0A195C3F4 B4KUX2 A0A067QRR4 A0A195ELE1 A0A151WHB5 A0A1W4VAD1 E0VU62 B4H8R1 Q29DN7 B4HIY7 A0A182GKZ5 A0A1Q3FF91 U5EVV3 B4QL00 B4J0Z7 A0A0J9RSJ4 Q16M97 A0A2P8YZ74 A0A3B0JYR9 A0A1Q3FBJ8 A0A1Q3FBJ4 A0A1Q3FAK2 A0A1Q3FAP7 B0W015 A0A3B0K1J8 A0A182GL46 Q9VSC9 F7E448 A0A0J7LAD4 A0A0M4ENB7 A0A336KZH6 A0A336N0I4 B4PCS1 B4LBZ7 K7IQ31 A0A232EQ44 T1GY78 D6WYN3 V5GRA1 A0A0L0C9U7 B3NF63 A0A2M4BCF2 A0A2M4BBD9 A0A2M4BBA1 A0A2M4BD80 A0A2M4ABB6 A0A2M4ABW3 W5JCZ2 A0A0T6B207 A0A182F361 A0A2M3Z5B1 A0A1B0D666 A0A1I8NB24 A0A182WFF8 Q7Q8G8
A0A212EQ76 A0A0L7LB05 A0A1B6CDV3 A0A1B6F1Z1 A0A1B6MA64 A0A1B6KMD1 A0A1B6DYW4 A0A0A9XSD2 A0A0A9XUH3 A0A0A9XZ38 A0A0K8TBS6 E2C738 A0A146LHA4 A0A0A9XZ45 A0A0A9XPA1 A0A0A9XSC8 A0A2S2QNI3 B4MXD2 A0A224XHV3 J9K213 A0A2A3EKI6 A0A069DZN1 A0A0P8YHW4 A0A2H8TF87 B3M431 A0A088APY5 A0A0C9R6L1 A0A195FRF2 A0A026VVX5 F4WC84 T1HMH1 A0A158N9Y4 B4IQA7 A0A195BK20 A0A0N0BCF2 A0A2S2PQQ2 A0A0L7R8G6 A0A195C3F4 B4KUX2 A0A067QRR4 A0A195ELE1 A0A151WHB5 A0A1W4VAD1 E0VU62 B4H8R1 Q29DN7 B4HIY7 A0A182GKZ5 A0A1Q3FF91 U5EVV3 B4QL00 B4J0Z7 A0A0J9RSJ4 Q16M97 A0A2P8YZ74 A0A3B0JYR9 A0A1Q3FBJ8 A0A1Q3FBJ4 A0A1Q3FAK2 A0A1Q3FAP7 B0W015 A0A3B0K1J8 A0A182GL46 Q9VSC9 F7E448 A0A0J7LAD4 A0A0M4ENB7 A0A336KZH6 A0A336N0I4 B4PCS1 B4LBZ7 K7IQ31 A0A232EQ44 T1GY78 D6WYN3 V5GRA1 A0A0L0C9U7 B3NF63 A0A2M4BCF2 A0A2M4BBD9 A0A2M4BBA1 A0A2M4BD80 A0A2M4ABB6 A0A2M4ABW3 W5JCZ2 A0A0T6B207 A0A182F361 A0A2M3Z5B1 A0A1B0D666 A0A1I8NB24 A0A182WFF8 Q7Q8G8
Ontologies
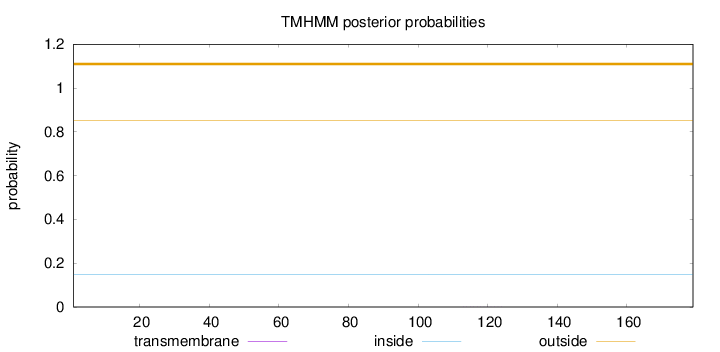
Topology
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00399
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.14828
outside
1 - 179
Population Genetic Test Statistics
Pi
286.218727
Theta
176.07614
Tajima's D
1.386879
CLR
0.192495
CSRT
0.75841207939603
Interpretation
Uncertain
