Gene
KWMTBOMO01325
Pre Gene Modal
BGIBMGA008882
Annotation
hypothetical_protein_KGM_18482_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.809
Sequence
CDS
ATGCAAACTTTGTATTTCACATTGGCACTTTTGAATGACTTATTTGGAAATAATGAGCCAACACCATCAGAGAAGCCTTTAATAAGACGCATAAAAGATATATTATTTAGTGCCTTGGCTTTTCCACTGTCTATGTTTGTTGGTATTACATTTTGGGGCATATACGCCATCGACAGAGAGCTCATTTTACCAAGAAGCTTAGACCCATATTTTCCTACATGGCTGAATCATGTAATGCACACAAACATAATGGCATTTATTCTAATTGATTTGTTAACATCCTTCAGAATGTATCCTAGGAAGAAAATTGGACTATCTGTACTTTGTACATATATGTTGTGTTACATGGTTTGGATTCATGTAATATATTTTAAAACTGGAAGTTGGGTGTATCCTATACTGAATATACTCAACTGGCCCCTAAGAGTAGTGTTCTATCTAGTCTGTCTTGGATTAGTTTGTAGTCTCTACAGCTTAGGCGAAAAATTTAACAAGTTGGTTTGGTCTAAAGAAGTAGAACAAACTGTTAAATCTGGAAAGAAAAAAGCAAAATAA
Protein
MQTLYFTLALLNDLFGNNEPTPSEKPLIRRIKDILFSALAFPLSMFVGITFWGIYAIDRELILPRSLDPYFPTWLNHVMHTNIMAFILIDLLTSFRMYPRKKIGLSVLCTYMLCYMVWIHVIYFKTGSWVYPILNILNWPLRVVFYLVCLGLVCSLYSLGEKFNKLVWSKEVEQTVKSGKKKAK
Summary
Uniprot
H9JH85
A0A2H1V657
A0A212EQ70
A0A0L7LAT4
A0A194RMK5
A0A2A4JX63
+ More
I4DNX5 S4PDG5 A0A2W1C1R7 D6WWA7 A0A067RJM4 A0A1W4WVR9 A0A1W4X6H3 A0A1W4X583 A0A2J7Q8Z3 A0A182HAH9 A0A1I8P877 A0A182P499 A0A1I8P8A7 A0A182GWI7 A0A023EL97 A0A1Y1LNK5 A0A182MZ94 A0A1Y1LPI5 A0A1Q3FGS6 B0W047 Q175G6 A0A1Q3FYK9 A0A182SRY6 A0A0C9REQ9 A0A034W8T5 N6TWE1 A0A182X5X5 A0A182V3W5 A0A182KN63 A0A182UHK6 A0A182IBK7 A0A1S4GYS9 Q7Q8H9 A0A182J862 A0A034WCY0 A0A1I8MHJ1 A0A1S4GYS4 T1PHV2 A0A182WFH3 A0A1Q3FH27 A0A0P9A870 B3MUK0 K7J3G6 A0A182LUY3 A0A084WFW8 A0A182RK64 A0A182QP29 A0A0N8NZJ7 A0A3F2YQ78 A0A0Q9X2B5 A0A182JRB5 A0A0M3QTA1 A0A1L8EHA6 A0A1L8EGS8 A0A0Q9WZ16 B4MWJ7 A0A2M4BX88 A0A1W4UQK1 A0A1W4V2H3 A0A1W4V2U5 A0A3F2YQ74 A0A182YN58 A0A2M4CLU5 W5JPW5 A0A0Q5W8I8 A0A1J1IP07 A0A0Q5W8L2 B3N7N2 A0A1A9ZFK5 A0A1J1ISG2 D3TLU3 A0A1B0AUL2 A0A1A9UTD2 A0A1A9XQI0 A0A0K8VIY7 A0A0R1DR51 A0A2M4CLR0 A1A6S8 Q9VPN0 A8DYS6 B4P2C0 A0A0J9TCF7 A0A0T6BDD8 A0A0K8VT93 A0A0J9QU08 Q9VPN1 B4ICU8 B4Q618 C6TP28 B4KHG8 A0A0Q9X5R3 A0A0R1DIH7 B4GJP9 Q29NW8
I4DNX5 S4PDG5 A0A2W1C1R7 D6WWA7 A0A067RJM4 A0A1W4WVR9 A0A1W4X6H3 A0A1W4X583 A0A2J7Q8Z3 A0A182HAH9 A0A1I8P877 A0A182P499 A0A1I8P8A7 A0A182GWI7 A0A023EL97 A0A1Y1LNK5 A0A182MZ94 A0A1Y1LPI5 A0A1Q3FGS6 B0W047 Q175G6 A0A1Q3FYK9 A0A182SRY6 A0A0C9REQ9 A0A034W8T5 N6TWE1 A0A182X5X5 A0A182V3W5 A0A182KN63 A0A182UHK6 A0A182IBK7 A0A1S4GYS9 Q7Q8H9 A0A182J862 A0A034WCY0 A0A1I8MHJ1 A0A1S4GYS4 T1PHV2 A0A182WFH3 A0A1Q3FH27 A0A0P9A870 B3MUK0 K7J3G6 A0A182LUY3 A0A084WFW8 A0A182RK64 A0A182QP29 A0A0N8NZJ7 A0A3F2YQ78 A0A0Q9X2B5 A0A182JRB5 A0A0M3QTA1 A0A1L8EHA6 A0A1L8EGS8 A0A0Q9WZ16 B4MWJ7 A0A2M4BX88 A0A1W4UQK1 A0A1W4V2H3 A0A1W4V2U5 A0A3F2YQ74 A0A182YN58 A0A2M4CLU5 W5JPW5 A0A0Q5W8I8 A0A1J1IP07 A0A0Q5W8L2 B3N7N2 A0A1A9ZFK5 A0A1J1ISG2 D3TLU3 A0A1B0AUL2 A0A1A9UTD2 A0A1A9XQI0 A0A0K8VIY7 A0A0R1DR51 A0A2M4CLR0 A1A6S8 Q9VPN0 A8DYS6 B4P2C0 A0A0J9TCF7 A0A0T6BDD8 A0A0K8VT93 A0A0J9QU08 Q9VPN1 B4ICU8 B4Q618 C6TP28 B4KHG8 A0A0Q9X5R3 A0A0R1DIH7 B4GJP9 Q29NW8
Pubmed
19121390
22118469
26227816
26354079
22651552
23622113
+ More
28756777 18362917 19820115 24845553 26483478 24945155 28004739 17510324 25348373 23537049 20966253 12364791 25315136 17994087 20075255 24438588 25244985 20920257 23761445 18057021 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085
28756777 18362917 19820115 24845553 26483478 24945155 28004739 17510324 25348373 23537049 20966253 12364791 25315136 17994087 20075255 24438588 25244985 20920257 23761445 18057021 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085
EMBL
BABH01012762
BABH01012763
BABH01012764
ODYU01000688
SOQ35882.1
AGBW02013301
+ More
OWR43635.1 JTDY01001898 KOB72587.1 KQ459989 KPJ18757.1 NWSH01000444 PCG76366.1 AK403186 BAM19615.1 GAIX01003691 JAA88869.1 KZ149894 PZC78906.1 KQ971361 EFA08684.2 KK852428 KDR24056.1 NEVH01016943 PNF25057.1 JXUM01031224 KQ560913 KXJ80373.1 JXUM01093221 KQ564042 KXJ72878.1 GAPW01003607 JAC09991.1 GEZM01054630 GEZM01054629 JAV73515.1 GEZM01054631 GEZM01054628 JAV73516.1 GFDL01008285 JAV26760.1 DS231816 EDS37619.1 CH477401 EAT41703.1 GFDL01002386 JAV32659.1 GBYB01006820 JAG76587.1 GAKP01006959 GAKP01006958 GAKP01006956 JAC51996.1 APGK01058767 KB741291 KB631759 ENN70612.1 ERL85849.1 APCN01000708 AAAB01008944 EAA10218.3 GAKP01006957 JAC51995.1 KA647705 AFP62334.1 GFDL01008187 JAV26858.1 CH902624 KPU74517.1 EDV33529.1 AXCM01003295 ATLV01023406 KE525343 KFB49112.1 AXCN02002069 KPU74516.1 CH963857 KRF98391.1 CP012523 ALC38553.1 GFDG01000826 JAV17973.1 GFDG01000954 JAV17845.1 KRF98392.1 EDW76138.1 GGFJ01008528 MBW57669.1 GGFL01002075 MBW66253.1 ADMH02000838 ETN64905.1 CH954177 KQS69920.1 CVRI01000055 CRL01466.1 KQS69919.1 KQS69921.1 EDV57208.1 CRL01465.1 EZ422395 ADD18671.1 JXJN01003674 GDHF01013458 JAI38856.1 CM000157 KRJ97030.1 GGFL01002076 MBW66254.1 BT029599 ABL75658.1 AE014134 AY060615 AAF51516.1 AAL28163.1 BT120126 ABV53595.1 ADB46407.1 EDW87116.1 CM002910 KMY87283.1 KMY87285.1 LJIG01002005 KRT84935.1 GDHF01010193 JAI42121.1 KMY87284.1 AAF51515.1 CH480829 EDW45374.1 CM000361 EDX03202.1 KMY87282.1 BT099514 ACU27356.1 CH933807 EDW11232.1 KRG02585.1 KRJ97031.1 CH479184 EDW36865.1 CH379058 EAL34525.2
OWR43635.1 JTDY01001898 KOB72587.1 KQ459989 KPJ18757.1 NWSH01000444 PCG76366.1 AK403186 BAM19615.1 GAIX01003691 JAA88869.1 KZ149894 PZC78906.1 KQ971361 EFA08684.2 KK852428 KDR24056.1 NEVH01016943 PNF25057.1 JXUM01031224 KQ560913 KXJ80373.1 JXUM01093221 KQ564042 KXJ72878.1 GAPW01003607 JAC09991.1 GEZM01054630 GEZM01054629 JAV73515.1 GEZM01054631 GEZM01054628 JAV73516.1 GFDL01008285 JAV26760.1 DS231816 EDS37619.1 CH477401 EAT41703.1 GFDL01002386 JAV32659.1 GBYB01006820 JAG76587.1 GAKP01006959 GAKP01006958 GAKP01006956 JAC51996.1 APGK01058767 KB741291 KB631759 ENN70612.1 ERL85849.1 APCN01000708 AAAB01008944 EAA10218.3 GAKP01006957 JAC51995.1 KA647705 AFP62334.1 GFDL01008187 JAV26858.1 CH902624 KPU74517.1 EDV33529.1 AXCM01003295 ATLV01023406 KE525343 KFB49112.1 AXCN02002069 KPU74516.1 CH963857 KRF98391.1 CP012523 ALC38553.1 GFDG01000826 JAV17973.1 GFDG01000954 JAV17845.1 KRF98392.1 EDW76138.1 GGFJ01008528 MBW57669.1 GGFL01002075 MBW66253.1 ADMH02000838 ETN64905.1 CH954177 KQS69920.1 CVRI01000055 CRL01466.1 KQS69919.1 KQS69921.1 EDV57208.1 CRL01465.1 EZ422395 ADD18671.1 JXJN01003674 GDHF01013458 JAI38856.1 CM000157 KRJ97030.1 GGFL01002076 MBW66254.1 BT029599 ABL75658.1 AE014134 AY060615 AAF51516.1 AAL28163.1 BT120126 ABV53595.1 ADB46407.1 EDW87116.1 CM002910 KMY87283.1 KMY87285.1 LJIG01002005 KRT84935.1 GDHF01010193 JAI42121.1 KMY87284.1 AAF51515.1 CH480829 EDW45374.1 CM000361 EDX03202.1 KMY87282.1 BT099514 ACU27356.1 CH933807 EDW11232.1 KRG02585.1 KRJ97031.1 CH479184 EDW36865.1 CH379058 EAL34525.2
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000218220
UP000007266
+ More
UP000027135 UP000192223 UP000235965 UP000069940 UP000249989 UP000095300 UP000075885 UP000075884 UP000002320 UP000008820 UP000075901 UP000019118 UP000030742 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075880 UP000095301 UP000075920 UP000007801 UP000002358 UP000075883 UP000030765 UP000075900 UP000075886 UP000069272 UP000007798 UP000075881 UP000092553 UP000192221 UP000076408 UP000000673 UP000008711 UP000183832 UP000092445 UP000092460 UP000078200 UP000092443 UP000002282 UP000000803 UP000001292 UP000000304 UP000009192 UP000008744 UP000001819
UP000027135 UP000192223 UP000235965 UP000069940 UP000249989 UP000095300 UP000075885 UP000075884 UP000002320 UP000008820 UP000075901 UP000019118 UP000030742 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075880 UP000095301 UP000075920 UP000007801 UP000002358 UP000075883 UP000030765 UP000075900 UP000075886 UP000069272 UP000007798 UP000075881 UP000092553 UP000192221 UP000076408 UP000000673 UP000008711 UP000183832 UP000092445 UP000092460 UP000078200 UP000092443 UP000002282 UP000000803 UP000001292 UP000000304 UP000009192 UP000008744 UP000001819
ProteinModelPortal
H9JH85
A0A2H1V657
A0A212EQ70
A0A0L7LAT4
A0A194RMK5
A0A2A4JX63
+ More
I4DNX5 S4PDG5 A0A2W1C1R7 D6WWA7 A0A067RJM4 A0A1W4WVR9 A0A1W4X6H3 A0A1W4X583 A0A2J7Q8Z3 A0A182HAH9 A0A1I8P877 A0A182P499 A0A1I8P8A7 A0A182GWI7 A0A023EL97 A0A1Y1LNK5 A0A182MZ94 A0A1Y1LPI5 A0A1Q3FGS6 B0W047 Q175G6 A0A1Q3FYK9 A0A182SRY6 A0A0C9REQ9 A0A034W8T5 N6TWE1 A0A182X5X5 A0A182V3W5 A0A182KN63 A0A182UHK6 A0A182IBK7 A0A1S4GYS9 Q7Q8H9 A0A182J862 A0A034WCY0 A0A1I8MHJ1 A0A1S4GYS4 T1PHV2 A0A182WFH3 A0A1Q3FH27 A0A0P9A870 B3MUK0 K7J3G6 A0A182LUY3 A0A084WFW8 A0A182RK64 A0A182QP29 A0A0N8NZJ7 A0A3F2YQ78 A0A0Q9X2B5 A0A182JRB5 A0A0M3QTA1 A0A1L8EHA6 A0A1L8EGS8 A0A0Q9WZ16 B4MWJ7 A0A2M4BX88 A0A1W4UQK1 A0A1W4V2H3 A0A1W4V2U5 A0A3F2YQ74 A0A182YN58 A0A2M4CLU5 W5JPW5 A0A0Q5W8I8 A0A1J1IP07 A0A0Q5W8L2 B3N7N2 A0A1A9ZFK5 A0A1J1ISG2 D3TLU3 A0A1B0AUL2 A0A1A9UTD2 A0A1A9XQI0 A0A0K8VIY7 A0A0R1DR51 A0A2M4CLR0 A1A6S8 Q9VPN0 A8DYS6 B4P2C0 A0A0J9TCF7 A0A0T6BDD8 A0A0K8VT93 A0A0J9QU08 Q9VPN1 B4ICU8 B4Q618 C6TP28 B4KHG8 A0A0Q9X5R3 A0A0R1DIH7 B4GJP9 Q29NW8
I4DNX5 S4PDG5 A0A2W1C1R7 D6WWA7 A0A067RJM4 A0A1W4WVR9 A0A1W4X6H3 A0A1W4X583 A0A2J7Q8Z3 A0A182HAH9 A0A1I8P877 A0A182P499 A0A1I8P8A7 A0A182GWI7 A0A023EL97 A0A1Y1LNK5 A0A182MZ94 A0A1Y1LPI5 A0A1Q3FGS6 B0W047 Q175G6 A0A1Q3FYK9 A0A182SRY6 A0A0C9REQ9 A0A034W8T5 N6TWE1 A0A182X5X5 A0A182V3W5 A0A182KN63 A0A182UHK6 A0A182IBK7 A0A1S4GYS9 Q7Q8H9 A0A182J862 A0A034WCY0 A0A1I8MHJ1 A0A1S4GYS4 T1PHV2 A0A182WFH3 A0A1Q3FH27 A0A0P9A870 B3MUK0 K7J3G6 A0A182LUY3 A0A084WFW8 A0A182RK64 A0A182QP29 A0A0N8NZJ7 A0A3F2YQ78 A0A0Q9X2B5 A0A182JRB5 A0A0M3QTA1 A0A1L8EHA6 A0A1L8EGS8 A0A0Q9WZ16 B4MWJ7 A0A2M4BX88 A0A1W4UQK1 A0A1W4V2H3 A0A1W4V2U5 A0A3F2YQ74 A0A182YN58 A0A2M4CLU5 W5JPW5 A0A0Q5W8I8 A0A1J1IP07 A0A0Q5W8L2 B3N7N2 A0A1A9ZFK5 A0A1J1ISG2 D3TLU3 A0A1B0AUL2 A0A1A9UTD2 A0A1A9XQI0 A0A0K8VIY7 A0A0R1DR51 A0A2M4CLR0 A1A6S8 Q9VPN0 A8DYS6 B4P2C0 A0A0J9TCF7 A0A0T6BDD8 A0A0K8VT93 A0A0J9QU08 Q9VPN1 B4ICU8 B4Q618 C6TP28 B4KHG8 A0A0Q9X5R3 A0A0R1DIH7 B4GJP9 Q29NW8
Ontologies
GO
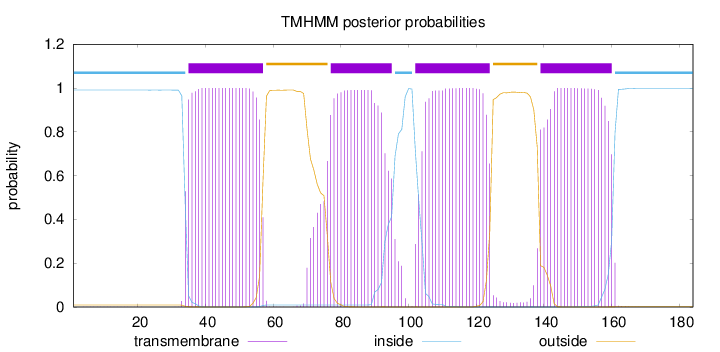
Topology
Length:
184
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.24154
Exp number, first 60 AAs:
22.69445
Total prob of N-in:
0.99057
POSSIBLE N-term signal
sequence
inside
1 - 34
TMhelix
35 - 57
outside
58 - 76
TMhelix
77 - 95
inside
96 - 101
TMhelix
102 - 124
outside
125 - 138
TMhelix
139 - 160
inside
161 - 184
Population Genetic Test Statistics
Pi
167.164299
Theta
164.714612
Tajima's D
0.241817
CLR
241.818763
CSRT
0.443177841107945
Interpretation
Uncertain
