Gene
KWMTBOMO01320 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008881
Annotation
ribosomal_protein_L11_[Bombyx_mori]
Full name
60S ribosomal protein L11
Location in the cell
Cytoplasmic Reliability : 2.301
Sequence
CDS
ATGGCGCGTGTACCACCGCCTGCGTTGAAGAAAGATAAAAAGGAGAAGAAGGTTCCGAAGGATAATTCTAAAAATGTAATGCGGAATCTTCATATCAGAAAGCTTTGCTTGAACATATGTGTTGGTGAATCCGGTGACAGGCTGACTCGTGCCGCCAAGGTGTTGGAGCAACTCACAGGACAACAGCCTGTATTTTCCAAGGCTAGGTATACAGTGCGGTCTTTTGGTATCCGTCGTAATGAAAAGATTGCTGTCCATTGTACAGTCCGAGGAGCTAAAGCAGAAGAAATCCTTGAGAGGGGTTTGAAAGTCAGAGAATATGAATTGCGGCGTGACAACTTCTCCGCCACGGGTAATTTTGGCTTCGGTATTCAAGAACACATTGACTTGGGTATCAAGTACGATCCCTCAATTGGAATTTATGGACTGGACTTTTACGTCGTACTTGGCCGGCCAGGTTTCAACGTAGCACACAGAAGGCGTAAGACAGGAAAAGTGGGATTTCCCCACCGCCTCACAAAGGAAGATGCTATGAAGTGGTTCCAACAGAAATATGATGGTATCATACTTAATAGCAAAGCAAAGTAA
Protein
MARVPPPALKKDKKEKKVPKDNSKNVMRNLHIRKLCLNICVGESGDRLTRAAKVLEQLTGQQPVFSKARYTVRSFGIRRNEKIAVHCTVRGAKAEEILERGLKVREYELRRDNFSATGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLGRPGFNVAHRRRKTGKVGFPHRLTKEDAMKWFQQKYDGIILNSKAK
Summary
Description
Component of the ribosome, a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. The small ribosomal subunit (SSU) binds messenger RNAs (mRNAs) and translates the encoded message by selecting cognate aminoacyl-transfer RNA (tRNA) molecules. The large subunit (LSU) contains the ribosomal catalytic site termed the peptidyl transferase center (PTC), which catalyzes the formation of peptide bonds, thereby polymerizing the amino acids delivered by tRNAs into a polypeptide chain. The nascent polypeptides leave the ribosome through a tunnel in the LSU and interact with protein factors that function in enzymatic processing, targeting, and the membrane insertion of nascent chains at the exit of the ribosomal tunnel.
Subunit
Component of the large ribosomal subunit (By similarity). Interacts with Fmr1 to form the RNA-induced silencing complex (RISC), a ribonucleoprotein (RNP) complex involved in translation regulation, other components of the complex are RpL5, Rm62, AGO2 and Dcr-1 (PubMed:12368261).
Similarity
Belongs to the universal ribosomal protein uL5 family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Nucleus
Reference proteome
Ribonucleoprotein
Ribosomal protein
RNA-binding
rRNA-binding
Feature
chain 60S ribosomal protein L11
Uniprot
Q5UAS8
H9JH84
I4DIR1
E7DZ01
S4PBC9
A0A194RMW5
+ More
A0A2W1BC20 A0A2A4JXE9 A0A2H1V4V5 A0A3G1T1J8 D1LYK2 G0ZEB8 D6WW88 Q4GXJ7 A0A0K8TR34 A0A034VVM3 A0A1A9WAD4 W8C038 A0A0C9QL70 A0A1Y1KT19 A0A0K2QK66 A0A1B0FL14 A0A1A9YJN7 A0A1A9UYE3 A0A1B0ARL8 A0A1A9ZRC6 J3JV68 T1PEX6 A0A0K8WIM5 A0A1B0ESU1 A0A0M4E7J9 B3MGG1 A0A1I8NMT8 A0A1L8EF71 A0A194PUA0 B4J622 A8CAG0 A0A1L8DRN6 A0A023EIH3 B4LNW5 A0A3B0J327 B5DUR4 B4GIM7 A0A1L8DS09 A0A0Q9WFK3 B0X194 B4KME8 A0A0Q9XIP5 A0A1Q3F9D3 A0A1W4WVQ0 Q5MIR0 A0A1L8DRW6 Q16KL2 A0A0L0C5G5 U5EXU7 B6U3W3 Q4GXJ8 A0A2M3YXE5 A0A2M3ZYT7 A0A182FKJ3 A0A182MVK4 A0A182YB37 A0A182WDH4 A0A0C9Q4H7 A0A182NZK8 A0A182QFW2 A0A171B8E9 A0A2M3ZF78 A0A182JJC1 A0A182N2D0 B6DDS7 A0A182JNJ3 Q06DH4 D1M7Y0 Q1HRK1 C4N198 B4MRQ7 A0A232ESU2 K7IRP4 A6YPT0 D3TS85 Q4GXJ6 A0A3B0JQN9 A0A0V0G2Q0 A0A069DQE7 A0A182VEB6 A0A182UBA1 A0A182X5L0 A0A182KL51 Q7QHF9 A0A182HWD9 A0A0B4LGZ5 B4PB83 B3NJV1 B4QE29 P46222 A0A026WUH2 A0A0L7QW18 V9IKH6 S4S5K2
A0A2W1BC20 A0A2A4JXE9 A0A2H1V4V5 A0A3G1T1J8 D1LYK2 G0ZEB8 D6WW88 Q4GXJ7 A0A0K8TR34 A0A034VVM3 A0A1A9WAD4 W8C038 A0A0C9QL70 A0A1Y1KT19 A0A0K2QK66 A0A1B0FL14 A0A1A9YJN7 A0A1A9UYE3 A0A1B0ARL8 A0A1A9ZRC6 J3JV68 T1PEX6 A0A0K8WIM5 A0A1B0ESU1 A0A0M4E7J9 B3MGG1 A0A1I8NMT8 A0A1L8EF71 A0A194PUA0 B4J622 A8CAG0 A0A1L8DRN6 A0A023EIH3 B4LNW5 A0A3B0J327 B5DUR4 B4GIM7 A0A1L8DS09 A0A0Q9WFK3 B0X194 B4KME8 A0A0Q9XIP5 A0A1Q3F9D3 A0A1W4WVQ0 Q5MIR0 A0A1L8DRW6 Q16KL2 A0A0L0C5G5 U5EXU7 B6U3W3 Q4GXJ8 A0A2M3YXE5 A0A2M3ZYT7 A0A182FKJ3 A0A182MVK4 A0A182YB37 A0A182WDH4 A0A0C9Q4H7 A0A182NZK8 A0A182QFW2 A0A171B8E9 A0A2M3ZF78 A0A182JJC1 A0A182N2D0 B6DDS7 A0A182JNJ3 Q06DH4 D1M7Y0 Q1HRK1 C4N198 B4MRQ7 A0A232ESU2 K7IRP4 A6YPT0 D3TS85 Q4GXJ6 A0A3B0JQN9 A0A0V0G2Q0 A0A069DQE7 A0A182VEB6 A0A182UBA1 A0A182X5L0 A0A182KL51 Q7QHF9 A0A182HWD9 A0A0B4LGZ5 B4PB83 B3NJV1 B4QE29 P46222 A0A026WUH2 A0A0L7QW18 V9IKH6 S4S5K2
Pubmed
19121390
22651552
23622113
26354079
28756777
18362917
+ More
19820115 26369729 25348373 24495485 28004739 26168724 22516182 23537049 25315136 17994087 17760985 24945155 26483478 15632085 17244540 17510324 26108605 18937034 25244985 19178717 20920257 23761445 17244545 17204158 19576987 28648823 20075255 18207082 20353571 26334808 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7893752 12537569 12368261 23636399 24508170
19820115 26369729 25348373 24495485 28004739 26168724 22516182 23537049 25315136 17994087 17760985 24945155 26483478 15632085 17244540 17510324 26108605 18937034 25244985 19178717 20920257 23761445 17244545 17204158 19576987 28648823 20075255 18207082 20353571 26334808 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7893752 12537569 12368261 23636399 24508170
EMBL
AY769280
AAV34822.1
BABH01012765
AK401179
BAM17801.1
HQ424704
+ More
ADT80648.1 GAIX01005972 JAA86588.1 KQ459989 KPJ18759.1 KZ150536 PZC70586.1 NWSH01000444 PCG76368.1 ODYU01000688 SOQ35880.1 MG992422 AXY94860.1 GU084276 ACY95309.1 JF264996 AEL28827.1 KQ971361 EFA08168.1 AM049026 CAJ17262.1 GDAI01001218 JAI16385.1 GAKP01013374 JAC45578.1 GAMC01001543 JAC05013.1 GBYB01001317 JAG71084.1 GEZM01074445 JAV64549.1 LC057254 BAS02547.1 CCAG010020342 JXJN01002477 APGK01035752 BT127135 KB740928 KB630163 KB630856 KB631023 AEE62097.1 ENN77943.1 ERL83441.1 ERL83907.1 ERL84014.1 KA647347 AFP61976.1 GDHF01001311 JAI51003.1 AJWK01005593 CP012524 ALC42691.1 CH902619 EDV35704.1 GFDG01001437 JAV17362.1 KQ459591 KPI96986.1 CH916367 EDW01880.1 EU045352 ABV44749.1 GFDF01005013 JAV09071.1 JXUM01135370 JXUM01135371 JXUM01135372 GAPW01004862 KQ568261 JAC08736.1 KXJ69073.1 CH940648 EDW61134.1 OUUW01000001 SPP75675.1 CH674335 EDY71633.1 CH479183 EDW36347.1 GFDF01004816 JAV09268.1 KRF79803.1 DS232256 EDS38543.1 CH933808 EDW09836.1 KRG04930.1 GFDL01010889 JAV24156.1 AY826150 AAV90722.1 GFDF01004994 JAV09090.1 CH477951 EAT34850.1 JRES01000890 KNC27525.1 GANO01002357 JAB57514.1 EU971928 ACG44046.1 AM049025 CAJ17261.1 GGFM01000186 MBW20937.1 GGFK01000350 MBW33671.1 AXCM01000599 GBYB01008973 JAG78740.1 AXCN02001727 GEMB01000148 JAS02965.1 GGFM01006390 MBW27141.1 EU934287 ADMH02002090 ACI30065.1 ETN59373.1 DQ910351 ABI83773.1 GU120404 ACY71254.1 DQ440093 ABF18126.1 EZ048871 ACN69163.1 CH963850 EDW74796.1 NNAY01002394 OXU21366.1 EF639091 ABR27976.1 EZ424287 ADD20563.1 AM049027 CAJ17263.1 SPP75676.1 GECL01003727 JAP02397.1 GBGD01003047 JAC85842.1 AAAB01008816 EAA05139.3 APCN01001485 AE013599 AHN56409.1 CM000158 EDW91496.1 CH954179 EDV55210.1 CM000362 CM002911 EDX07816.1 KMY95118.1 U15643 AY094790 KK107102 EZA59603.1 KQ414716 KOC62828.1 JR049563 AEY61001.1 HQ828077 HQ828081 JN699055 AET36542.1 AET74064.1
ADT80648.1 GAIX01005972 JAA86588.1 KQ459989 KPJ18759.1 KZ150536 PZC70586.1 NWSH01000444 PCG76368.1 ODYU01000688 SOQ35880.1 MG992422 AXY94860.1 GU084276 ACY95309.1 JF264996 AEL28827.1 KQ971361 EFA08168.1 AM049026 CAJ17262.1 GDAI01001218 JAI16385.1 GAKP01013374 JAC45578.1 GAMC01001543 JAC05013.1 GBYB01001317 JAG71084.1 GEZM01074445 JAV64549.1 LC057254 BAS02547.1 CCAG010020342 JXJN01002477 APGK01035752 BT127135 KB740928 KB630163 KB630856 KB631023 AEE62097.1 ENN77943.1 ERL83441.1 ERL83907.1 ERL84014.1 KA647347 AFP61976.1 GDHF01001311 JAI51003.1 AJWK01005593 CP012524 ALC42691.1 CH902619 EDV35704.1 GFDG01001437 JAV17362.1 KQ459591 KPI96986.1 CH916367 EDW01880.1 EU045352 ABV44749.1 GFDF01005013 JAV09071.1 JXUM01135370 JXUM01135371 JXUM01135372 GAPW01004862 KQ568261 JAC08736.1 KXJ69073.1 CH940648 EDW61134.1 OUUW01000001 SPP75675.1 CH674335 EDY71633.1 CH479183 EDW36347.1 GFDF01004816 JAV09268.1 KRF79803.1 DS232256 EDS38543.1 CH933808 EDW09836.1 KRG04930.1 GFDL01010889 JAV24156.1 AY826150 AAV90722.1 GFDF01004994 JAV09090.1 CH477951 EAT34850.1 JRES01000890 KNC27525.1 GANO01002357 JAB57514.1 EU971928 ACG44046.1 AM049025 CAJ17261.1 GGFM01000186 MBW20937.1 GGFK01000350 MBW33671.1 AXCM01000599 GBYB01008973 JAG78740.1 AXCN02001727 GEMB01000148 JAS02965.1 GGFM01006390 MBW27141.1 EU934287 ADMH02002090 ACI30065.1 ETN59373.1 DQ910351 ABI83773.1 GU120404 ACY71254.1 DQ440093 ABF18126.1 EZ048871 ACN69163.1 CH963850 EDW74796.1 NNAY01002394 OXU21366.1 EF639091 ABR27976.1 EZ424287 ADD20563.1 AM049027 CAJ17263.1 SPP75676.1 GECL01003727 JAP02397.1 GBGD01003047 JAC85842.1 AAAB01008816 EAA05139.3 APCN01001485 AE013599 AHN56409.1 CM000158 EDW91496.1 CH954179 EDV55210.1 CM000362 CM002911 EDX07816.1 KMY95118.1 U15643 AY094790 KK107102 EZA59603.1 KQ414716 KOC62828.1 JR049563 AEY61001.1 HQ828077 HQ828081 JN699055 AET36542.1 AET74064.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007266
UP000091820
UP000092444
+ More
UP000092443 UP000078200 UP000092460 UP000092445 UP000019118 UP000030742 UP000095301 UP000092461 UP000092553 UP000007801 UP000095300 UP000053268 UP000001070 UP000069940 UP000249989 UP000008792 UP000268350 UP000001819 UP000008744 UP000002320 UP000009192 UP000192223 UP000008820 UP000037069 UP000069272 UP000075883 UP000076408 UP000075920 UP000075885 UP000075886 UP000075880 UP000075884 UP000000673 UP000075881 UP000075900 UP000007798 UP000215335 UP000002358 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000803 UP000002282 UP000008711 UP000000304 UP000053097 UP000053825
UP000092443 UP000078200 UP000092460 UP000092445 UP000019118 UP000030742 UP000095301 UP000092461 UP000092553 UP000007801 UP000095300 UP000053268 UP000001070 UP000069940 UP000249989 UP000008792 UP000268350 UP000001819 UP000008744 UP000002320 UP000009192 UP000192223 UP000008820 UP000037069 UP000069272 UP000075883 UP000076408 UP000075920 UP000075885 UP000075886 UP000075880 UP000075884 UP000000673 UP000075881 UP000075900 UP000007798 UP000215335 UP000002358 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000803 UP000002282 UP000008711 UP000000304 UP000053097 UP000053825
Interpro
SUPFAM
SSF55282
SSF55282
Gene 3D
ProteinModelPortal
Q5UAS8
H9JH84
I4DIR1
E7DZ01
S4PBC9
A0A194RMW5
+ More
A0A2W1BC20 A0A2A4JXE9 A0A2H1V4V5 A0A3G1T1J8 D1LYK2 G0ZEB8 D6WW88 Q4GXJ7 A0A0K8TR34 A0A034VVM3 A0A1A9WAD4 W8C038 A0A0C9QL70 A0A1Y1KT19 A0A0K2QK66 A0A1B0FL14 A0A1A9YJN7 A0A1A9UYE3 A0A1B0ARL8 A0A1A9ZRC6 J3JV68 T1PEX6 A0A0K8WIM5 A0A1B0ESU1 A0A0M4E7J9 B3MGG1 A0A1I8NMT8 A0A1L8EF71 A0A194PUA0 B4J622 A8CAG0 A0A1L8DRN6 A0A023EIH3 B4LNW5 A0A3B0J327 B5DUR4 B4GIM7 A0A1L8DS09 A0A0Q9WFK3 B0X194 B4KME8 A0A0Q9XIP5 A0A1Q3F9D3 A0A1W4WVQ0 Q5MIR0 A0A1L8DRW6 Q16KL2 A0A0L0C5G5 U5EXU7 B6U3W3 Q4GXJ8 A0A2M3YXE5 A0A2M3ZYT7 A0A182FKJ3 A0A182MVK4 A0A182YB37 A0A182WDH4 A0A0C9Q4H7 A0A182NZK8 A0A182QFW2 A0A171B8E9 A0A2M3ZF78 A0A182JJC1 A0A182N2D0 B6DDS7 A0A182JNJ3 Q06DH4 D1M7Y0 Q1HRK1 C4N198 B4MRQ7 A0A232ESU2 K7IRP4 A6YPT0 D3TS85 Q4GXJ6 A0A3B0JQN9 A0A0V0G2Q0 A0A069DQE7 A0A182VEB6 A0A182UBA1 A0A182X5L0 A0A182KL51 Q7QHF9 A0A182HWD9 A0A0B4LGZ5 B4PB83 B3NJV1 B4QE29 P46222 A0A026WUH2 A0A0L7QW18 V9IKH6 S4S5K2
A0A2W1BC20 A0A2A4JXE9 A0A2H1V4V5 A0A3G1T1J8 D1LYK2 G0ZEB8 D6WW88 Q4GXJ7 A0A0K8TR34 A0A034VVM3 A0A1A9WAD4 W8C038 A0A0C9QL70 A0A1Y1KT19 A0A0K2QK66 A0A1B0FL14 A0A1A9YJN7 A0A1A9UYE3 A0A1B0ARL8 A0A1A9ZRC6 J3JV68 T1PEX6 A0A0K8WIM5 A0A1B0ESU1 A0A0M4E7J9 B3MGG1 A0A1I8NMT8 A0A1L8EF71 A0A194PUA0 B4J622 A8CAG0 A0A1L8DRN6 A0A023EIH3 B4LNW5 A0A3B0J327 B5DUR4 B4GIM7 A0A1L8DS09 A0A0Q9WFK3 B0X194 B4KME8 A0A0Q9XIP5 A0A1Q3F9D3 A0A1W4WVQ0 Q5MIR0 A0A1L8DRW6 Q16KL2 A0A0L0C5G5 U5EXU7 B6U3W3 Q4GXJ8 A0A2M3YXE5 A0A2M3ZYT7 A0A182FKJ3 A0A182MVK4 A0A182YB37 A0A182WDH4 A0A0C9Q4H7 A0A182NZK8 A0A182QFW2 A0A171B8E9 A0A2M3ZF78 A0A182JJC1 A0A182N2D0 B6DDS7 A0A182JNJ3 Q06DH4 D1M7Y0 Q1HRK1 C4N198 B4MRQ7 A0A232ESU2 K7IRP4 A6YPT0 D3TS85 Q4GXJ6 A0A3B0JQN9 A0A0V0G2Q0 A0A069DQE7 A0A182VEB6 A0A182UBA1 A0A182X5L0 A0A182KL51 Q7QHF9 A0A182HWD9 A0A0B4LGZ5 B4PB83 B3NJV1 B4QE29 P46222 A0A026WUH2 A0A0L7QW18 V9IKH6 S4S5K2
PDB
4V6W
E-value=9.95339e-89,
Score=829
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
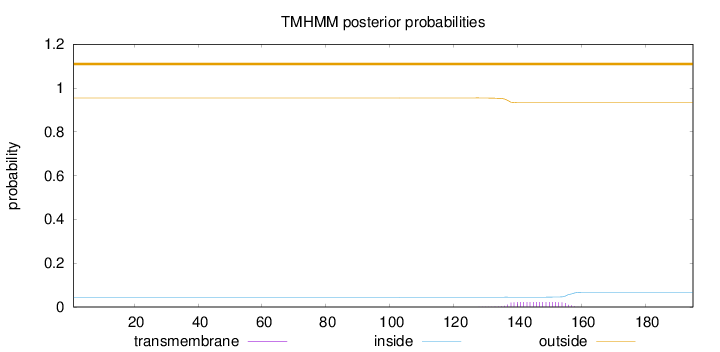
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04507
outside
1 - 195
Population Genetic Test Statistics
Pi
215.922165
Theta
172.615262
Tajima's D
0.767341
CLR
0.142026
CSRT
0.589770511474426
Interpretation
Uncertain
