Gene
KWMTBOMO01315 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000596
Annotation
vacuolar_ATP_synthase_subunit_H_[Bombyx_mori]
Full name
V-type proton ATPase subunit H
Alternative Name
Vacuolar proton pump subunit H
Vacuolar proton pump subunit SFD
Vacuolar proton pump subunit SFD
Location in the cell
Cytoplasmic Reliability : 2.03
Sequence
CDS
ATGGCTAATGTTAGCGAGGAAAATGTTAGTCAGTTAATTCCAACGCTAGGTGACGAAAAAATCGACATGATTGCGGCCACCAGCGTCCTACAGATCCGGGCAAGTGAGATACGCCAAACTCGGATTAACTGGCAGTCATATCTGCAGTCGCAGATGATCACTCAGCGCGATCACGATTTTATCGTGAACTTAGACCAGCGCGGCCAAAAAGATCTGCCTGACAAGAATCCTGATGCCTGTGCCGAAGTCTTTCTCAACCTCCTGACGCATATCAGCAAGGACCACACCATTCAATACATTCTTGTGCTCATTGATGACATTCTTTCTGAAGATAAATCGAGGGTGAAGATATTTCGTGAGACGAAATTCTCTGGCAACGTTTGGCAGCCCTTCCTGAATTTGCTGAACCGTCAGGATGAATTTGTCCAGCACATGACTGCTCGCATCATTGCCAAGCTAGCTTGCTGGCATCCGCAGCTGATGGACAAAAGTGATTTGCACTTCTATCTCTCCTGGCTTAAAGATCAACTCAAGACCAATAACAACGACTATATCCAGTCTGTGGCCCGGTGTCTGCAGATGATGCTGCGTATCGATGAATACCGCTTTGCTTTCCTCTCTGTCGATGGTATATCCACCTTACTGTCGATTCTGGCCTCCAGAGTGAACTTCCAGGTTCAATACCAACTTGTATTTTGTCTCTGGGTTTTGACATTCAATCCCCTATTGGCTGAAAAAATGAACAAGTTCAATGTTATTCCAATCTTAGCTGATATCCTTAGCGACTCCGTTAAAGAGAAGGTCACGCGTATTGTGCTCGCAGTGTTCAGGAATTTAATTGAAAAACCTGAAGATCAACAGGTTGCCAAAGAGCACTGCATTGCCATGGTACAGTGCAAGGTACTCAAGCAACTGTCAATCCTTGAACAGAAGCGTTCTGATGATGAGGATATTATGAATGATGTGGAATATTTGAATGAACGTCTACAAGCTTCCGTACAGGATCTTAGTTCTTTTGATCAATATGCTACTGAAGTAAAGAGTGGCCGCCTGGAATGGTCGCCGGTACACAAATCGGCCAAGTTCTGGCGCGAAAACGCGGCTCGTTTGAACGAACGTGGCCAAGAACTGCTCCGCACCCTGGTGCACTTGCTGGAGAAGAGCCGCGACCCTGTCGTACTCGCCGTCGCTTGCTATGACATCGGAGAATATGTGCGCCACTATCCGCGCGGCAAACACATCATCGAACAACTTGGTGGTAAACAACGTGTCATGTACCTCCTGAGTCACGACGATCCGAATGTACGTTACGAAGCCTTGCTCGCCGTGCAGAAACTTATGGTTCACAACTGGGAATATCTCGGCAAGCAACTGGAGAAGGAACAAATCGACAAACAGGCTGGCACTGTGGTTGGAGCTAAGGCATAA
Protein
MANVSEENVSQLIPTLGDEKIDMIAATSVLQIRASEIRQTRINWQSYLQSQMITQRDHDFIVNLDQRGQKDLPDKNPDACAEVFLNLLTHISKDHTIQYILVLIDDILSEDKSRVKIFRETKFSGNVWQPFLNLLNRQDEFVQHMTARIIAKLACWHPQLMDKSDLHFYLSWLKDQLKTNNNDYIQSVARCLQMMLRIDEYRFAFLSVDGISTLLSILASRVNFQVQYQLVFCLWVLTFNPLLAEKMNKFNVIPILADILSDSVKEKVTRIVLAVFRNLIEKPEDQQVAKEHCIAMVQCKVLKQLSILEQKRSDDEDIMNDVEYLNERLQASVQDLSSFDQYATEVKSGRLEWSPVHKSAKFWRENAARLNERGQELLRTLVHLLEKSRDPVVLAVACYDIGEYVRHYPRGKHIIEQLGGKQRVMYLLSHDDPNVRYEALLAVQKLMVHNWEYLGKQLEKEQIDKQAGTVVGAKA
Summary
Description
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit H activates ATPase activity of the enzyme and couples ATPase activity to proton flow. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system.
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit H activates the ATPase activity of the enzyme and couples ATPase activity to proton flow. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system (By similarity).
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit H activates the ATPase activity of the enzyme and couples ATPase activity to proton flow. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system (By similarity).
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex.
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Similarity
Belongs to the V-ATPase H subunit family.
Keywords
Hydrogen ion transport
Ion transport
Transport
Alternative splicing
Complete proteome
Reference proteome
Feature
chain V-type proton ATPase subunit H
splice variant In isoform B.
splice variant In isoform B.
Uniprot
Q1HPJ9
Q9U5N0
H9ITL6
A0A2A4J7U6
I4DJP3
S4PX42
+ More
A0A1E1WF42 A0A0D3L6K4 A0A0D3L6K9 A0A194RLQ0 A0A194PW52 A0A0C9RCZ0 A0A2J7RRL5 A0A023ZZ51 V5I8V7 A0A2D1QUD7 A0A026WVU6 K7IWJ4 A0A084VPJ7 Q0IF50 A0A023ETY0 A0A1Y1LRC7 A0A1L8DF92 A0A2M4CTQ0 A0A2M3Z0F2 A0A1W4WVG5 U5EWK6 A0A2J7RRL0 A0A2M4BM69 A0A1L8DFF0 A0A158P3I8 B0W5Q3 A0A1L8DFK7 A0A2M4A477 A0A182RGC9 A0A1Q3FJ75 A0A182PA86 V9IK54 A0A2C9H1C2 T1DQR3 A0A0J7KQU2 A0A069DV46 A0A1B6K9Z3 A0A1B6FZX2 A0A161M902 R4G5L0 A0A224XBP5 J3JVP9 A0A1I8PQW3 A0A151HYZ3 A0A0V0G4Q3 A0A023F9H7 E0VQC4 A0A2R7WGG3 A0A0P4VM08 A0A2A3EB40 A0A1L8EER0 A0A0A9XMX6 A0A0K8U503 A0A034W025 R4WIU5 A0A0C9RPY1 A0A0A1WQ73 T1PB95 A0A1W4VXR0 A0A1I8MD39 A0A0N8NZJ1 A0A0R3NSM8 A0A0Q9W948 U4UPV4 M9PDK5 Q9V3J1 A0A0J9TPK3 A0A0Q9XH85 A0A0Q9WQ72 A0A1B6DSM1 A0A3B0KB61 A0A151JXJ8 A0A0A9YKV4 A0A0Q5W2M8 A0A0R1DRB4 D3TS59 A0A2S2R3U4 A0A1I9WLJ4 A0A2H8TY66 A0A1S3CTK5 A0A1B6BXI2 X2F5F3 Q9V3J1-3 A0A0J9R3A5 J9JPW6 T1JCY2 A0A0R1DQ25 A0A0Q5VN59 A0A2S2PDQ8 A0A2R5L869 A0A293N2U5 A0A1L8DF87 A0A162CNC8 A0A0P6B3U8
A0A1E1WF42 A0A0D3L6K4 A0A0D3L6K9 A0A194RLQ0 A0A194PW52 A0A0C9RCZ0 A0A2J7RRL5 A0A023ZZ51 V5I8V7 A0A2D1QUD7 A0A026WVU6 K7IWJ4 A0A084VPJ7 Q0IF50 A0A023ETY0 A0A1Y1LRC7 A0A1L8DF92 A0A2M4CTQ0 A0A2M3Z0F2 A0A1W4WVG5 U5EWK6 A0A2J7RRL0 A0A2M4BM69 A0A1L8DFF0 A0A158P3I8 B0W5Q3 A0A1L8DFK7 A0A2M4A477 A0A182RGC9 A0A1Q3FJ75 A0A182PA86 V9IK54 A0A2C9H1C2 T1DQR3 A0A0J7KQU2 A0A069DV46 A0A1B6K9Z3 A0A1B6FZX2 A0A161M902 R4G5L0 A0A224XBP5 J3JVP9 A0A1I8PQW3 A0A151HYZ3 A0A0V0G4Q3 A0A023F9H7 E0VQC4 A0A2R7WGG3 A0A0P4VM08 A0A2A3EB40 A0A1L8EER0 A0A0A9XMX6 A0A0K8U503 A0A034W025 R4WIU5 A0A0C9RPY1 A0A0A1WQ73 T1PB95 A0A1W4VXR0 A0A1I8MD39 A0A0N8NZJ1 A0A0R3NSM8 A0A0Q9W948 U4UPV4 M9PDK5 Q9V3J1 A0A0J9TPK3 A0A0Q9XH85 A0A0Q9WQ72 A0A1B6DSM1 A0A3B0KB61 A0A151JXJ8 A0A0A9YKV4 A0A0Q5W2M8 A0A0R1DRB4 D3TS59 A0A2S2R3U4 A0A1I9WLJ4 A0A2H8TY66 A0A1S3CTK5 A0A1B6BXI2 X2F5F3 Q9V3J1-3 A0A0J9R3A5 J9JPW6 T1JCY2 A0A0R1DQ25 A0A0Q5VN59 A0A2S2PDQ8 A0A2R5L869 A0A293N2U5 A0A1L8DF87 A0A162CNC8 A0A0P6B3U8
Pubmed
11030595
19121390
22651552
23622113
26354079
25863352
+ More
24508170 20075255 24438588 17510324 24945155 28004739 21347285 12364791 26334808 22516182 25474469 20566863 27129103 25401762 26823975 25348373 23691247 25830018 25315136 17994087 15632085 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 17550304 20353571 27538518
24508170 20075255 24438588 17510324 24945155 28004739 21347285 12364791 26334808 22516182 25474469 20566863 27129103 25401762 26823975 25348373 23691247 25830018 25315136 17994087 15632085 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 17550304 20353571 27538518
EMBL
DQ443403
ABF51492.1
AJ249389
BABH01006739
NWSH01002535
PCG68041.1
+ More
AK401511 BAM18133.1 GAIX01005693 JAA86867.1 GDQN01005446 JAT85608.1 KC683729 AHF70968.1 KC683730 AHF70969.1 KQ459989 KPJ18763.1 KQ459591 KPI96989.1 GBYB01014454 JAG84221.1 NEVH01000599 PNF43479.1 KJ476830 AHY84718.1 GALX01004015 JAB64451.1 KY285046 ATP16149.1 KK107088 EZA59801.1 ATLV01015007 KE524999 KFB39891.1 CH477394 EAT41889.1 GAPW01001247 JAC12351.1 GEZM01054530 JAV73577.1 GFDF01008968 JAV05116.1 GGFL01004471 MBW68649.1 GGFM01001256 MBW22007.1 GANO01001374 JAB58497.1 PNF43478.1 GGFJ01004991 MBW54132.1 GFDF01008969 JAV05115.1 ADTU01008088 ADTU01008089 DS231844 EDS35706.1 GFDF01008970 JAV05114.1 GGFK01002293 MBW35614.1 GFDL01007471 JAV27574.1 JR049161 AEY60881.1 AAAB01008839 GAMD01001010 JAB00581.1 LBMM01004251 KMQ92661.1 GBGD01001312 JAC87577.1 GEBQ01031711 JAT08266.1 GECZ01014030 JAS55739.1 GEMB01004302 JAR98970.1 ACPB03017385 GAHY01000440 JAA77070.1 GFTR01006579 JAW09847.1 BT127317 AEE62279.1 KQ976710 KYM77039.1 GECL01003460 JAP02664.1 GBBI01000672 JAC18040.1 DS235418 EEB15580.1 KK854773 PTY18589.1 GDKW01001054 JAI55541.1 KZ288311 PBC28512.1 GFDG01001665 JAV17134.1 GBHO01025134 GDHC01007726 JAG18470.1 JAQ10903.1 GDHF01030859 JAI21455.1 GAKP01011809 JAC47143.1 AK417400 BAN20615.1 GBYB01009241 JAG79008.1 GBXI01013290 JAD01002.1 KA645997 AFP60626.1 CH902624 KPU74468.1 CH379060 KRT04025.1 CH940649 KRF81293.1 KB632408 ERL95137.1 AE014134 AGB93045.1 AF159457 AY052122 CM002910 KMY90555.1 CH933807 KRG03601.1 CH963857 KRF98314.1 GEDC01008627 JAS28671.1 OUUW01000006 SPP82291.1 KQ981604 KYN39720.1 GBHO01011871 GDHC01000236 JAG31733.1 JAQ18393.1 CH954179 KQS62959.1 CM000158 KRJ99069.1 CCAG010018508 EZ424261 ADD20537.1 GGMS01015466 MBY84669.1 KU932378 APA34014.1 GFXV01006936 MBW18741.1 GEDC01031306 JAS05992.1 KJ018752 AHM92104.1 KMY90553.1 ABLF02006768 JH432085 KRJ99070.1 KQS62957.1 GGMR01014971 MBY27590.1 GGLE01001534 MBY05660.1 GFWV01022666 MAA47393.1 GFDF01008986 JAV05098.1 LRGB01000930 KZS14926.1 GDIP01034194 JAM69521.1
AK401511 BAM18133.1 GAIX01005693 JAA86867.1 GDQN01005446 JAT85608.1 KC683729 AHF70968.1 KC683730 AHF70969.1 KQ459989 KPJ18763.1 KQ459591 KPI96989.1 GBYB01014454 JAG84221.1 NEVH01000599 PNF43479.1 KJ476830 AHY84718.1 GALX01004015 JAB64451.1 KY285046 ATP16149.1 KK107088 EZA59801.1 ATLV01015007 KE524999 KFB39891.1 CH477394 EAT41889.1 GAPW01001247 JAC12351.1 GEZM01054530 JAV73577.1 GFDF01008968 JAV05116.1 GGFL01004471 MBW68649.1 GGFM01001256 MBW22007.1 GANO01001374 JAB58497.1 PNF43478.1 GGFJ01004991 MBW54132.1 GFDF01008969 JAV05115.1 ADTU01008088 ADTU01008089 DS231844 EDS35706.1 GFDF01008970 JAV05114.1 GGFK01002293 MBW35614.1 GFDL01007471 JAV27574.1 JR049161 AEY60881.1 AAAB01008839 GAMD01001010 JAB00581.1 LBMM01004251 KMQ92661.1 GBGD01001312 JAC87577.1 GEBQ01031711 JAT08266.1 GECZ01014030 JAS55739.1 GEMB01004302 JAR98970.1 ACPB03017385 GAHY01000440 JAA77070.1 GFTR01006579 JAW09847.1 BT127317 AEE62279.1 KQ976710 KYM77039.1 GECL01003460 JAP02664.1 GBBI01000672 JAC18040.1 DS235418 EEB15580.1 KK854773 PTY18589.1 GDKW01001054 JAI55541.1 KZ288311 PBC28512.1 GFDG01001665 JAV17134.1 GBHO01025134 GDHC01007726 JAG18470.1 JAQ10903.1 GDHF01030859 JAI21455.1 GAKP01011809 JAC47143.1 AK417400 BAN20615.1 GBYB01009241 JAG79008.1 GBXI01013290 JAD01002.1 KA645997 AFP60626.1 CH902624 KPU74468.1 CH379060 KRT04025.1 CH940649 KRF81293.1 KB632408 ERL95137.1 AE014134 AGB93045.1 AF159457 AY052122 CM002910 KMY90555.1 CH933807 KRG03601.1 CH963857 KRF98314.1 GEDC01008627 JAS28671.1 OUUW01000006 SPP82291.1 KQ981604 KYN39720.1 GBHO01011871 GDHC01000236 JAG31733.1 JAQ18393.1 CH954179 KQS62959.1 CM000158 KRJ99069.1 CCAG010018508 EZ424261 ADD20537.1 GGMS01015466 MBY84669.1 KU932378 APA34014.1 GFXV01006936 MBW18741.1 GEDC01031306 JAS05992.1 KJ018752 AHM92104.1 KMY90553.1 ABLF02006768 JH432085 KRJ99070.1 KQS62957.1 GGMR01014971 MBY27590.1 GGLE01001534 MBY05660.1 GFWV01022666 MAA47393.1 GFDF01008986 JAV05098.1 LRGB01000930 KZS14926.1 GDIP01034194 JAM69521.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000235965
UP000053097
+ More
UP000002358 UP000030765 UP000008820 UP000192223 UP000005205 UP000002320 UP000075900 UP000075885 UP000036403 UP000015103 UP000095300 UP000078540 UP000009046 UP000242457 UP000192221 UP000095301 UP000007801 UP000001819 UP000008792 UP000030742 UP000000803 UP000009192 UP000007798 UP000268350 UP000078541 UP000008711 UP000002282 UP000092444 UP000079169 UP000007819 UP000076858
UP000002358 UP000030765 UP000008820 UP000192223 UP000005205 UP000002320 UP000075900 UP000075885 UP000036403 UP000015103 UP000095300 UP000078540 UP000009046 UP000242457 UP000192221 UP000095301 UP000007801 UP000001819 UP000008792 UP000030742 UP000000803 UP000009192 UP000007798 UP000268350 UP000078541 UP000008711 UP000002282 UP000092444 UP000079169 UP000007819 UP000076858
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
Q1HPJ9
Q9U5N0
H9ITL6
A0A2A4J7U6
I4DJP3
S4PX42
+ More
A0A1E1WF42 A0A0D3L6K4 A0A0D3L6K9 A0A194RLQ0 A0A194PW52 A0A0C9RCZ0 A0A2J7RRL5 A0A023ZZ51 V5I8V7 A0A2D1QUD7 A0A026WVU6 K7IWJ4 A0A084VPJ7 Q0IF50 A0A023ETY0 A0A1Y1LRC7 A0A1L8DF92 A0A2M4CTQ0 A0A2M3Z0F2 A0A1W4WVG5 U5EWK6 A0A2J7RRL0 A0A2M4BM69 A0A1L8DFF0 A0A158P3I8 B0W5Q3 A0A1L8DFK7 A0A2M4A477 A0A182RGC9 A0A1Q3FJ75 A0A182PA86 V9IK54 A0A2C9H1C2 T1DQR3 A0A0J7KQU2 A0A069DV46 A0A1B6K9Z3 A0A1B6FZX2 A0A161M902 R4G5L0 A0A224XBP5 J3JVP9 A0A1I8PQW3 A0A151HYZ3 A0A0V0G4Q3 A0A023F9H7 E0VQC4 A0A2R7WGG3 A0A0P4VM08 A0A2A3EB40 A0A1L8EER0 A0A0A9XMX6 A0A0K8U503 A0A034W025 R4WIU5 A0A0C9RPY1 A0A0A1WQ73 T1PB95 A0A1W4VXR0 A0A1I8MD39 A0A0N8NZJ1 A0A0R3NSM8 A0A0Q9W948 U4UPV4 M9PDK5 Q9V3J1 A0A0J9TPK3 A0A0Q9XH85 A0A0Q9WQ72 A0A1B6DSM1 A0A3B0KB61 A0A151JXJ8 A0A0A9YKV4 A0A0Q5W2M8 A0A0R1DRB4 D3TS59 A0A2S2R3U4 A0A1I9WLJ4 A0A2H8TY66 A0A1S3CTK5 A0A1B6BXI2 X2F5F3 Q9V3J1-3 A0A0J9R3A5 J9JPW6 T1JCY2 A0A0R1DQ25 A0A0Q5VN59 A0A2S2PDQ8 A0A2R5L869 A0A293N2U5 A0A1L8DF87 A0A162CNC8 A0A0P6B3U8
A0A1E1WF42 A0A0D3L6K4 A0A0D3L6K9 A0A194RLQ0 A0A194PW52 A0A0C9RCZ0 A0A2J7RRL5 A0A023ZZ51 V5I8V7 A0A2D1QUD7 A0A026WVU6 K7IWJ4 A0A084VPJ7 Q0IF50 A0A023ETY0 A0A1Y1LRC7 A0A1L8DF92 A0A2M4CTQ0 A0A2M3Z0F2 A0A1W4WVG5 U5EWK6 A0A2J7RRL0 A0A2M4BM69 A0A1L8DFF0 A0A158P3I8 B0W5Q3 A0A1L8DFK7 A0A2M4A477 A0A182RGC9 A0A1Q3FJ75 A0A182PA86 V9IK54 A0A2C9H1C2 T1DQR3 A0A0J7KQU2 A0A069DV46 A0A1B6K9Z3 A0A1B6FZX2 A0A161M902 R4G5L0 A0A224XBP5 J3JVP9 A0A1I8PQW3 A0A151HYZ3 A0A0V0G4Q3 A0A023F9H7 E0VQC4 A0A2R7WGG3 A0A0P4VM08 A0A2A3EB40 A0A1L8EER0 A0A0A9XMX6 A0A0K8U503 A0A034W025 R4WIU5 A0A0C9RPY1 A0A0A1WQ73 T1PB95 A0A1W4VXR0 A0A1I8MD39 A0A0N8NZJ1 A0A0R3NSM8 A0A0Q9W948 U4UPV4 M9PDK5 Q9V3J1 A0A0J9TPK3 A0A0Q9XH85 A0A0Q9WQ72 A0A1B6DSM1 A0A3B0KB61 A0A151JXJ8 A0A0A9YKV4 A0A0Q5W2M8 A0A0R1DRB4 D3TS59 A0A2S2R3U4 A0A1I9WLJ4 A0A2H8TY66 A0A1S3CTK5 A0A1B6BXI2 X2F5F3 Q9V3J1-3 A0A0J9R3A5 J9JPW6 T1JCY2 A0A0R1DQ25 A0A0Q5VN59 A0A2S2PDQ8 A0A2R5L869 A0A293N2U5 A0A1L8DF87 A0A162CNC8 A0A0P6B3U8
PDB
1HO8
E-value=5.47021e-24,
Score=276
Ontologies
KEGG
PATHWAY
GO
PANTHER
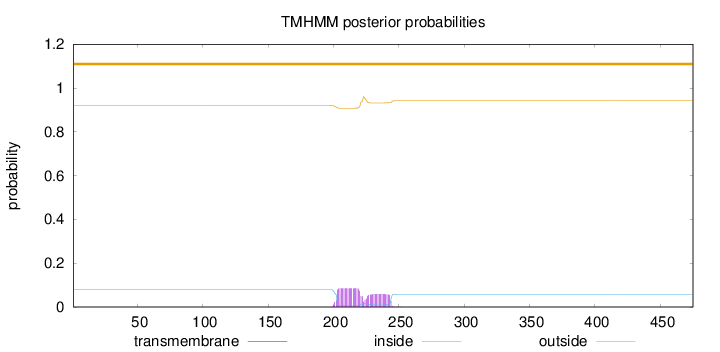
Topology
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.83499
Exp number, first 60 AAs:
0.00108
Total prob of N-in:
0.07901
outside
1 - 475
Population Genetic Test Statistics
Pi
297.924289
Theta
215.377214
Tajima's D
0.774624
CLR
0.018961
CSRT
0.593520323983801
Interpretation
Uncertain
