Gene
KWMTBOMO01309
Pre Gene Modal
BGIBMGA008876
Annotation
PREDICTED:_monocarboxylate_transporter_4-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.623
Sequence
CDS
ATGGAGGCAGGCTGGTGGTCACCGTCGGCCTCGCCGCCGCCGCCGCCCCGCGCCCCCCCGCCTCGCTGCCGCTGCCGTCACCCCTTCTCTACTAGCTACACCTGCTTGCCGCATTACGATGGATGGCCAGACGGGGAAGAGGAATCCACAGGAGGAGCGCTTCGGGAACGCGAGCTGCGGTCACTGCACCGCACCATCCCCGCGCTGCGGAGAAGAGAACTCGACACCCGGGCTATCAAGCACCACTTCTACCCTGAGGGAGGATGGGGATGGATCGTTTGTGGAGCGGCCTTTCTTGCACACTTATTGTCAACCGGACTGCAGCTGGCTTACGGAGCGCTACACGTTTACGCTTTGAAACACCTTGGACCAACCGCAGATCATGCTGTGTGGGCCGGAGCTATTTGCGTGGGAGTGTCCCGTGCTGCCGGTGCTCTTGTAGCGGGACGGAGACGGTCACCAAGGCTCGTGGCTTTACTCGGGGGCTTACTGCTGCCTTTGGCCTGCCTCTTCACATCCTTCGCGACACAGCTACATCAAACGTTACTAAGCTACGGTGTTGTACTTGGCATTGGATGCGGCTTAGTAAGAGAAGCAGCAGGCCTAGTACTTGGAGCATATTTTCGTAGAAGAAGACAATTTGTAGAACTAGTGGCTCATGCTGGCGGTGGTGTAGGCATTGCTCTGTTTAGCGTTGCATACAAAGAAGCAGTTGGAAAGCTTGGTTGGCGATTAGGATTGCAAGCGGTCACGGGAGTCTTAGTCTTGGCATTTTTTCTAAGTGCTGTTTACCGCAGTGCCTCTTTATACCATCCTCAACGCAGAGCTATATTACATTTAAAAAATCAACGTCGAAAGGTGAAGGAAAAGAAGGGTATAAAACGACCTCCACTTATTGATCTAAGCCCACTTCAATCTCGACCCGCGAAAGTTTTGTTGTTGGCCGCCGGATTAGCGGCTTTCGGACTTTATACCCCAGTATTTTTTCTGGCTTTACAGGGATTTCAGGAGGGCTTAGAGGAGAGTGCTTTGGTGCTGCTACAGACGTTTTTGGGATTCGCAGCGGTGCTCGGGTGTGCAGGCTTTGGACTCGTTCTTGTACGACCTTCGGCGCAATGTCTGGTCTCTAAGCAGTACTTATGTCAAACAGCGATGCTCGGTATAGGCATCTCAATGCTCGCTCTGAGCAGCGTCGAAGGTTATCATGGCTATGTCCTTTTCGCGTGGATGTACGGGCTATGTCTCGGTGGATATCTGTATTCTCTCAAAATGCTGACAATGGAAAGGGTTCGTGGTCGACACTTCACTAAAGTTTGGGGTTACGTTCAAGGAGCCGAAGCAATCCCTGTTATAGTAGGCGTTCCCGTAACAGGCTACATAAACCAGCAAGCTCCCAGGGCGGGATTCTATTTTTCGACGGCGGCAACATTGGCGGGCGCGTTCCTTTTGTTTTTCGTTGGATTTTCGAAACGCGAGCCGGAACCACCTCCGCCCGCTGCCGCTCCCTCCATATCGGAGTCCTGTCGGTGTACGAGTCCCCTGCGGTGCGGGGCAGGGGGCGCGTGGTGCGCGTGCGGTGCTGGAGGCGCAGCTGCATACTGTGCGTGGCCGGGACCAGCGTGCGGTGCGCGGCTACCCAAATCATTGTCGTACGCAGCACCTTTGGATAGGGCATGCTGCTCGGCTACGCACTGCCCGGACTGTTACCGTCAAACGGTGCCATTGCGTTCATCCCGCAGCGTCCCAGAGGGCCTGGCGACGCGCACATCATCGTGGGGTCGCGCCGGCTCTTGTCGCGGTGCGTGCCGGCGACGTGAACATCATCTTATAGAACAAATCACTACATCTGTTTGA
Protein
MEAGWWSPSASPPPPPRAPPPRCRCRHPFSTSYTCLPHYDGWPDGEEESTGGALRERELRSLHRTIPALRRRELDTRAIKHHFYPEGGWGWIVCGAAFLAHLLSTGLQLAYGALHVYALKHLGPTADHAVWAGAICVGVSRAAGALVAGRRRSPRLVALLGGLLLPLACLFTSFATQLHQTLLSYGVVLGIGCGLVREAAGLVLGAYFRRRRQFVELVAHAGGGVGIALFSVAYKEAVGKLGWRLGLQAVTGVLVLAFFLSAVYRSASLYHPQRRAILHLKNQRRKVKEKKGIKRPPLIDLSPLQSRPAKVLLLAAGLAAFGLYTPVFFLALQGFQEGLEESALVLLQTFLGFAAVLGCAGFGLVLVRPSAQCLVSKQYLCQTAMLGIGISMLALSSVEGYHGYVLFAWMYGLCLGGYLYSLKMLTMERVRGRHFTKVWGYVQGAEAIPVIVGVPVTGYINQQAPRAGFYFSTAATLAGAFLLFFVGFSKREPEPPPPAAAPSISESCRCTSPLRCGAGGAWCACGAGGAAAYCAWPGPACGARLPKSLSYAAPLDRACCSATHCPDCYRQTVPLRSSRSVPEGLATRTSSWGRAGSCRGACRRREHHLIEQITTSV
Summary
Uniprot
A0A2W1BJH6
A0A2A4JQP1
A0A194PW57
A0A194RN63
A0A2H1WST4
A0A212EPZ2
+ More
A0A2J7QHL5 D6WWS1 A0A1Y1MZR3 E0VX85 A0A0L7LDA7 A0A3L8D3F5 A0A158NQL8 A0A026W850 A0A067QUN6 A0A151XAX4 A0A195ERP4 A0A2A3EIM0 A0A195BRZ6 A0A2P8ZF61 A0A2M4BE40 A0A1S4G7T5 A0A0P4Y3H2 A0A0N8CJD3 A0A0P5SDU3 A0A1S4H0Z9 A0A0P5IHZ6 A0A0P6EGZ4 A0A0P5LJ80 A0A0P5L5B1 A0A0P5SK05 A0A0P5YUW9 A0A0P5W7W0 A0A0P6I4P3 A0A0P5L4B9 A0A0P5YV29 A0A0P5SLU2 A0A0P5ZAS3 A0A0P5I925 A0A0P6IAZ2 A0A182QZX5 A0A0P5VFV8 A0A0P4XH08 A0A0P4XKN3 A0A0P6BC52 A0A0P4ZGW9 B4LRR5 B4KJY5 B4JQJ4 A0A3B0K0C5 B4MVB1 A0A0M3QTH0 A0A3B0JAQ0 A0A0J9QW66 Q9VQU7 A0A1W4V6Z9 B4NYF1 B3MP65 B4Q9Q1 B3N375 B4G9C4 B4I333 Q29MF2 A0A1W4XIT9 A0A0L0C1V9 W8C166
A0A2J7QHL5 D6WWS1 A0A1Y1MZR3 E0VX85 A0A0L7LDA7 A0A3L8D3F5 A0A158NQL8 A0A026W850 A0A067QUN6 A0A151XAX4 A0A195ERP4 A0A2A3EIM0 A0A195BRZ6 A0A2P8ZF61 A0A2M4BE40 A0A1S4G7T5 A0A0P4Y3H2 A0A0N8CJD3 A0A0P5SDU3 A0A1S4H0Z9 A0A0P5IHZ6 A0A0P6EGZ4 A0A0P5LJ80 A0A0P5L5B1 A0A0P5SK05 A0A0P5YUW9 A0A0P5W7W0 A0A0P6I4P3 A0A0P5L4B9 A0A0P5YV29 A0A0P5SLU2 A0A0P5ZAS3 A0A0P5I925 A0A0P6IAZ2 A0A182QZX5 A0A0P5VFV8 A0A0P4XH08 A0A0P4XKN3 A0A0P6BC52 A0A0P4ZGW9 B4LRR5 B4KJY5 B4JQJ4 A0A3B0K0C5 B4MVB1 A0A0M3QTH0 A0A3B0JAQ0 A0A0J9QW66 Q9VQU7 A0A1W4V6Z9 B4NYF1 B3MP65 B4Q9Q1 B3N375 B4G9C4 B4I333 Q29MF2 A0A1W4XIT9 A0A0L0C1V9 W8C166
Pubmed
EMBL
KZ150048
PZC74461.1
NWSH01000882
PCG73730.1
KQ459591
KPI96994.1
+ More
KQ459989 KPJ18770.1 ODYU01010803 SOQ56130.1 AGBW02013349 OWR43549.1 NEVH01013973 PNF28097.1 KQ971361 EFA08751.1 GEZM01018746 JAV90040.1 DS235829 EEB17991.1 JTDY01001684 KOB73166.1 QOIP01000014 RLU14920.1 ADTU01023392 ADTU01023393 ADTU01023394 ADTU01023395 ADTU01023396 KK107405 EZA51194.1 KK852931 KDR13636.1 KQ982335 KYQ57533.1 KQ982021 KYN30572.1 KZ288232 PBC31548.1 KQ976419 KYM89284.1 PYGN01000076 PSN55128.1 GGFJ01002178 MBW51319.1 GDIP01232967 JAI90434.1 GDIP01125983 JAL77731.1 GDIP01141378 JAL62336.1 AAAB01008984 GDIP01238373 GDIQ01216094 JAI85028.1 JAK35631.1 GDIQ01063489 GDIQ01061486 JAN31248.1 GDIQ01177301 JAK74424.1 GDIQ01175392 JAK76333.1 GDIP01142477 JAL61237.1 GDIP01053043 JAM50672.1 GDIP01090027 JAM13688.1 GDIQ01035257 JAN59480.1 GDIQ01180596 JAK71129.1 GDIP01053042 JAM50673.1 GDIP01138174 JAL65540.1 GDIP01046798 JAM56917.1 GDIQ01217101 JAK34624.1 GDIQ01007120 JAN87617.1 AXCN02000838 GDIP01115620 JAL88094.1 GDIP01241260 JAI82141.1 GDIP01241259 JAI82142.1 GDIP01016715 JAM87000.1 GDIP01214059 JAJ09343.1 CH940649 EDW64667.2 CH933807 EDW12588.2 CH916372 EDV99174.1 OUUW01000004 SPP79066.1 CH963857 EDW76456.2 CP012523 ALC38869.1 SPP79065.1 CM002910 KMY87969.1 AE014134 AAF51067.2 AGB92538.1 CM000157 EDW87584.2 CH902620 EDV31231.2 CM000361 EDX03662.1 CH954177 EDV57674.2 CH479180 EDW28954.1 CH480820 EDW54178.1 CH379060 EAL33742.3 JRES01001119 KNC25389.1 GAMC01003702 JAC02854.1
KQ459989 KPJ18770.1 ODYU01010803 SOQ56130.1 AGBW02013349 OWR43549.1 NEVH01013973 PNF28097.1 KQ971361 EFA08751.1 GEZM01018746 JAV90040.1 DS235829 EEB17991.1 JTDY01001684 KOB73166.1 QOIP01000014 RLU14920.1 ADTU01023392 ADTU01023393 ADTU01023394 ADTU01023395 ADTU01023396 KK107405 EZA51194.1 KK852931 KDR13636.1 KQ982335 KYQ57533.1 KQ982021 KYN30572.1 KZ288232 PBC31548.1 KQ976419 KYM89284.1 PYGN01000076 PSN55128.1 GGFJ01002178 MBW51319.1 GDIP01232967 JAI90434.1 GDIP01125983 JAL77731.1 GDIP01141378 JAL62336.1 AAAB01008984 GDIP01238373 GDIQ01216094 JAI85028.1 JAK35631.1 GDIQ01063489 GDIQ01061486 JAN31248.1 GDIQ01177301 JAK74424.1 GDIQ01175392 JAK76333.1 GDIP01142477 JAL61237.1 GDIP01053043 JAM50672.1 GDIP01090027 JAM13688.1 GDIQ01035257 JAN59480.1 GDIQ01180596 JAK71129.1 GDIP01053042 JAM50673.1 GDIP01138174 JAL65540.1 GDIP01046798 JAM56917.1 GDIQ01217101 JAK34624.1 GDIQ01007120 JAN87617.1 AXCN02000838 GDIP01115620 JAL88094.1 GDIP01241260 JAI82141.1 GDIP01241259 JAI82142.1 GDIP01016715 JAM87000.1 GDIP01214059 JAJ09343.1 CH940649 EDW64667.2 CH933807 EDW12588.2 CH916372 EDV99174.1 OUUW01000004 SPP79066.1 CH963857 EDW76456.2 CP012523 ALC38869.1 SPP79065.1 CM002910 KMY87969.1 AE014134 AAF51067.2 AGB92538.1 CM000157 EDW87584.2 CH902620 EDV31231.2 CM000361 EDX03662.1 CH954177 EDV57674.2 CH479180 EDW28954.1 CH480820 EDW54178.1 CH379060 EAL33742.3 JRES01001119 KNC25389.1 GAMC01003702 JAC02854.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
UP000007266
+ More
UP000009046 UP000037510 UP000279307 UP000005205 UP000053097 UP000027135 UP000075809 UP000078541 UP000242457 UP000078540 UP000245037 UP000075886 UP000008792 UP000009192 UP000001070 UP000268350 UP000007798 UP000092553 UP000000803 UP000192221 UP000002282 UP000007801 UP000000304 UP000008711 UP000008744 UP000001292 UP000001819 UP000192223 UP000037069
UP000009046 UP000037510 UP000279307 UP000005205 UP000053097 UP000027135 UP000075809 UP000078541 UP000242457 UP000078540 UP000245037 UP000075886 UP000008792 UP000009192 UP000001070 UP000268350 UP000007798 UP000092553 UP000000803 UP000192221 UP000002282 UP000007801 UP000000304 UP000008711 UP000008744 UP000001292 UP000001819 UP000192223 UP000037069
PRIDE
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2W1BJH6
A0A2A4JQP1
A0A194PW57
A0A194RN63
A0A2H1WST4
A0A212EPZ2
+ More
A0A2J7QHL5 D6WWS1 A0A1Y1MZR3 E0VX85 A0A0L7LDA7 A0A3L8D3F5 A0A158NQL8 A0A026W850 A0A067QUN6 A0A151XAX4 A0A195ERP4 A0A2A3EIM0 A0A195BRZ6 A0A2P8ZF61 A0A2M4BE40 A0A1S4G7T5 A0A0P4Y3H2 A0A0N8CJD3 A0A0P5SDU3 A0A1S4H0Z9 A0A0P5IHZ6 A0A0P6EGZ4 A0A0P5LJ80 A0A0P5L5B1 A0A0P5SK05 A0A0P5YUW9 A0A0P5W7W0 A0A0P6I4P3 A0A0P5L4B9 A0A0P5YV29 A0A0P5SLU2 A0A0P5ZAS3 A0A0P5I925 A0A0P6IAZ2 A0A182QZX5 A0A0P5VFV8 A0A0P4XH08 A0A0P4XKN3 A0A0P6BC52 A0A0P4ZGW9 B4LRR5 B4KJY5 B4JQJ4 A0A3B0K0C5 B4MVB1 A0A0M3QTH0 A0A3B0JAQ0 A0A0J9QW66 Q9VQU7 A0A1W4V6Z9 B4NYF1 B3MP65 B4Q9Q1 B3N375 B4G9C4 B4I333 Q29MF2 A0A1W4XIT9 A0A0L0C1V9 W8C166
A0A2J7QHL5 D6WWS1 A0A1Y1MZR3 E0VX85 A0A0L7LDA7 A0A3L8D3F5 A0A158NQL8 A0A026W850 A0A067QUN6 A0A151XAX4 A0A195ERP4 A0A2A3EIM0 A0A195BRZ6 A0A2P8ZF61 A0A2M4BE40 A0A1S4G7T5 A0A0P4Y3H2 A0A0N8CJD3 A0A0P5SDU3 A0A1S4H0Z9 A0A0P5IHZ6 A0A0P6EGZ4 A0A0P5LJ80 A0A0P5L5B1 A0A0P5SK05 A0A0P5YUW9 A0A0P5W7W0 A0A0P6I4P3 A0A0P5L4B9 A0A0P5YV29 A0A0P5SLU2 A0A0P5ZAS3 A0A0P5I925 A0A0P6IAZ2 A0A182QZX5 A0A0P5VFV8 A0A0P4XH08 A0A0P4XKN3 A0A0P6BC52 A0A0P4ZGW9 B4LRR5 B4KJY5 B4JQJ4 A0A3B0K0C5 B4MVB1 A0A0M3QTH0 A0A3B0JAQ0 A0A0J9QW66 Q9VQU7 A0A1W4V6Z9 B4NYF1 B3MP65 B4Q9Q1 B3N375 B4G9C4 B4I333 Q29MF2 A0A1W4XIT9 A0A0L0C1V9 W8C166
Ontologies
GO
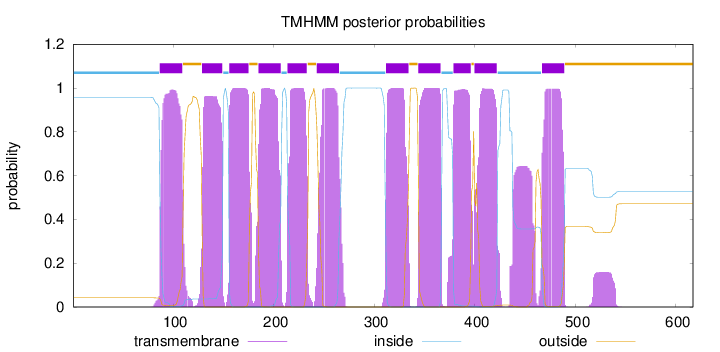
Topology
Length:
617
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
253.63661
Exp number, first 60 AAs:
0
Total prob of N-in:
0.95824
inside
1 - 86
TMhelix
87 - 109
outside
110 - 128
TMhelix
129 - 149
inside
150 - 155
TMhelix
156 - 175
outside
176 - 184
TMhelix
185 - 207
inside
208 - 213
TMhelix
214 - 233
outside
234 - 242
TMhelix
243 - 265
inside
266 - 311
TMhelix
312 - 334
outside
335 - 343
TMhelix
344 - 366
inside
367 - 378
TMhelix
379 - 396
outside
397 - 399
TMhelix
400 - 422
inside
423 - 466
TMhelix
467 - 489
outside
490 - 617
Population Genetic Test Statistics
Pi
219.003253
Theta
154.125443
Tajima's D
1.121304
CLR
0.259076
CSRT
0.694365281735913
Interpretation
Uncertain
