Gene
KWMTBOMO01303
Pre Gene Modal
BGIBMGA009013
Annotation
Sentrin-specific_protease_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.613 PlasmaMembrane Reliability : 1.085
Sequence
CDS
ATGGCAAAAGCATCTGTTGTGTTAAGTTTTCATGATATATTGCTTCATCGGTCTGATATAGAATTACTGGAAGGTCCGCATTGGTTGAATGATACGATAATATCATTTTACTTTGAATATCTCGAAAAAGTTGTGTTCAGTAAAGAGAGTGATCTATTATTTGTGTCTCCAGAGGTAACACAATGTATAAAAATGGTTCAAACAGAAGAAATAAAGACTTTTCTAGAACCGCTTGATGTGAACTTGAAGAAGTTTGTGTTCTTTGCCTTGAATGACAATAATACTCCAGATATGGCAGGTGGCTCCCATTGGAGCCTACTTGTTTATTCAAGGCCAGAGAACTGCTTCTTCCATTTAGATTCTTCACAAGCAACAAATCATGATGTTGCTTGGGAATTTGCAAGCCATATAATGTCATATCTTGCCAAAGGGGGTTCCATTAACTTTGTCGACAAGGAATGCATTCAGCAAAGTAACGGCTATGACTGTGGAATTCATGTAATAATCAATGCAGAGAGGCTTGCTGAATACTCTTGTGATGTTACCATGTGTTAA
Protein
MAKASVVLSFHDILLHRSDIELLEGPHWLNDTIISFYFEYLEKVVFSKESDLLFVSPEVTQCIKMVQTEEIKTFLEPLDVNLKKFVFFALNDNNTPDMAGGSHWSLLVYSRPENCFFHLDSSQATNHDVAWEFASHIMSYLAKGGSINFVDKECIQQSNGYDCGIHVIINAERLAEYSCDVTMC
Summary
Uniprot
H9JHL5
A0A0L7KVL8
A0A2A4J671
S4PXU9
A0A2W1BLE0
A0A2H1VC33
+ More
A0A023EJW9 A0A182G440 A0A1Y1LFY8 E0VQE1 A0A2M4BZE8 A0A182FLI6 A0A2J7Q959 Q179X8 W5JVX8 A0A1W4X9H4 A0A1B0CV02 A0A182K2V5 A0A2M4AT16 A0A182Y6Y5 A0A182N3E4 A0A2P8ZEI2 A0A182P0U1 A0A182RLL2 A0A182X2H2 Q7QHR4 A0A182HY69 A0A182TY24 A0A182WFM6 A0A182LHC6 A0A084VR21 A0A1Q3F837 B0XF62 U5EZA9 A0A182MDY6 D6WYW7 A0A182IXQ4 A0A182QIW3 A0A182VKD7 J3JVT9 A0A1B0GQF7 A0A336M6D3 A0A034VRG2 A0A0A1XB95 A0A0K8UQ10 A0A232ET42 K7IXL0 W8BF28 U4TUK6 N6SZB8 A0A210QSH5 A0A0Q5VZI0 B4HP91 B4QCT0 Q7JUX2 A0A0P4VSF4 A0A0R1DQB9 E1JH43 A0A0J9RBU6 R4WSK8 B3NS38 A0A0L0BUX2 T1PG56 B4P5D4 C0PV27 A0A2R7VX62 A0A1B0G7S1 A0A1A9UH13 A0A1A9ZI31 A0A1B6HQW8 A0A0P9BX41 A0A1I8Q4S2 A0A1A9WPP4 A0A1A9Y0P7 A0A1B0B4D4 A0A0Q9W2Y3 A0A0Q9XAN6 A0A3B0JNJ5 A0A3B0JVP2 B4LJQ3 B4MY68 B4KT88 B4JW50 B3MCA9 T1GDP9 A0A1B6LMV8 A0A067RJR1 A0A3B0J227 A0A1W4UJH0 A0A0M4EVF7 A0A1B6GQ56 A0A0R3NR06 A0A1W4U6P0 Q292E1 B4GDN5 A0A3N0XTI3 B4KT90
A0A023EJW9 A0A182G440 A0A1Y1LFY8 E0VQE1 A0A2M4BZE8 A0A182FLI6 A0A2J7Q959 Q179X8 W5JVX8 A0A1W4X9H4 A0A1B0CV02 A0A182K2V5 A0A2M4AT16 A0A182Y6Y5 A0A182N3E4 A0A2P8ZEI2 A0A182P0U1 A0A182RLL2 A0A182X2H2 Q7QHR4 A0A182HY69 A0A182TY24 A0A182WFM6 A0A182LHC6 A0A084VR21 A0A1Q3F837 B0XF62 U5EZA9 A0A182MDY6 D6WYW7 A0A182IXQ4 A0A182QIW3 A0A182VKD7 J3JVT9 A0A1B0GQF7 A0A336M6D3 A0A034VRG2 A0A0A1XB95 A0A0K8UQ10 A0A232ET42 K7IXL0 W8BF28 U4TUK6 N6SZB8 A0A210QSH5 A0A0Q5VZI0 B4HP91 B4QCT0 Q7JUX2 A0A0P4VSF4 A0A0R1DQB9 E1JH43 A0A0J9RBU6 R4WSK8 B3NS38 A0A0L0BUX2 T1PG56 B4P5D4 C0PV27 A0A2R7VX62 A0A1B0G7S1 A0A1A9UH13 A0A1A9ZI31 A0A1B6HQW8 A0A0P9BX41 A0A1I8Q4S2 A0A1A9WPP4 A0A1A9Y0P7 A0A1B0B4D4 A0A0Q9W2Y3 A0A0Q9XAN6 A0A3B0JNJ5 A0A3B0JVP2 B4LJQ3 B4MY68 B4KT88 B4JW50 B3MCA9 T1GDP9 A0A1B6LMV8 A0A067RJR1 A0A3B0J227 A0A1W4UJH0 A0A0M4EVF7 A0A1B6GQ56 A0A0R3NR06 A0A1W4U6P0 Q292E1 B4GDN5 A0A3N0XTI3 B4KT90
Pubmed
19121390
26227816
23622113
28756777
24945155
26483478
+ More
28004739 20566863 17510324 20920257 23761445 25244985 29403074 12364791 14747013 17210077 20966253 24438588 18362917 19820115 22516182 25348373 25830018 28648823 20075255 24495485 23537049 28812685 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23691247 26108605 25315136 24845553 15632085
28004739 20566863 17510324 20920257 23761445 25244985 29403074 12364791 14747013 17210077 20966253 24438588 18362917 19820115 22516182 25348373 25830018 28648823 20075255 24495485 23537049 28812685 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23691247 26108605 25315136 24845553 15632085
EMBL
BABH01012799
JTDY01005185
KOB67278.1
NWSH01002950
PCG67236.1
GAIX01004408
+ More
JAA88152.1 KZ150048 PZC74464.1 ODYU01001762 SOQ38409.1 JXUM01032192 GAPW01004238 KQ560948 JAC09360.1 KXJ80235.1 JXUM01006941 KQ560227 KXJ83738.1 GEZM01056866 JAV72582.1 DS235418 EEB15597.1 GGFJ01009251 MBW58392.1 NEVH01016943 PNF25127.1 CH477343 EAT43057.1 ADMH02000250 ETN67245.1 AJWK01029929 GGFK01010615 MBW43936.1 PYGN01000079 PSN54914.1 AAAB01008816 EAA05220.3 APCN01004496 ATLV01015425 KE525013 KFB40415.1 GFDL01011372 JAV23673.1 DS232900 EDS26475.1 GANO01001124 JAB58747.1 AXCM01001995 KQ971363 EFA07853.1 AXCN02001111 BT127357 AEE62319.1 AJVK01007205 UFQS01000198 UFQT01000198 SSX01208.1 SSX21588.1 GAKP01014814 GAKP01014812 GAKP01014810 GAKP01014808 GAKP01014806 JAC44138.1 GBXI01005643 GBXI01003529 JAD08649.1 JAD10763.1 GDHF01025622 GDHF01024676 GDHF01023550 GDHF01011666 JAI26692.1 JAI27638.1 JAI28764.1 JAI40648.1 NNAY01002333 OXU21525.1 AAZX01000028 GAMC01009279 GAMC01009278 JAB97277.1 KB631579 ERL84447.1 APGK01058731 KB741291 ENN70573.1 NEDP02002179 OWF51662.1 CH954179 KQS62846.1 CH480816 EDW47540.1 CM000362 CM002911 EDX06741.1 KMY93165.1 AE013599 AY119635 AAF58530.1 AAM50289.1 GDRN01100882 GDRN01100881 JAI58470.1 CM000158 KRJ99466.1 ACZ94401.1 KMY93164.1 AK417692 BAN20907.1 EDV56340.2 JRES01001422 KNC23024.1 KA647797 AFP62426.1 EDW90796.2 BT072883 ACN67107.1 KK854143 PTY12052.1 CCAG010016893 GECU01030638 JAS77068.1 CH902619 KPU76085.1 JXJN01008264 CH940648 KRF79482.1 CH933808 KRG05341.1 OUUW01000001 SPP75139.1 SPP75138.1 EDW60562.2 CH963894 EDW77057.1 EDW10600.2 CH916375 EDV98188.1 EDV36209.2 CAQQ02023992 GEBQ01014912 JAT25065.1 KK852428 KDR24047.1 SPP75137.1 CP012524 ALC41850.1 GECZ01010494 GECZ01005230 JAS59275.1 JAS64539.1 CM000071 KRT01695.1 EAL24921.3 CH479181 EDW31692.1 RJVU01061435 ROJ36095.1 EDW10602.2
JAA88152.1 KZ150048 PZC74464.1 ODYU01001762 SOQ38409.1 JXUM01032192 GAPW01004238 KQ560948 JAC09360.1 KXJ80235.1 JXUM01006941 KQ560227 KXJ83738.1 GEZM01056866 JAV72582.1 DS235418 EEB15597.1 GGFJ01009251 MBW58392.1 NEVH01016943 PNF25127.1 CH477343 EAT43057.1 ADMH02000250 ETN67245.1 AJWK01029929 GGFK01010615 MBW43936.1 PYGN01000079 PSN54914.1 AAAB01008816 EAA05220.3 APCN01004496 ATLV01015425 KE525013 KFB40415.1 GFDL01011372 JAV23673.1 DS232900 EDS26475.1 GANO01001124 JAB58747.1 AXCM01001995 KQ971363 EFA07853.1 AXCN02001111 BT127357 AEE62319.1 AJVK01007205 UFQS01000198 UFQT01000198 SSX01208.1 SSX21588.1 GAKP01014814 GAKP01014812 GAKP01014810 GAKP01014808 GAKP01014806 JAC44138.1 GBXI01005643 GBXI01003529 JAD08649.1 JAD10763.1 GDHF01025622 GDHF01024676 GDHF01023550 GDHF01011666 JAI26692.1 JAI27638.1 JAI28764.1 JAI40648.1 NNAY01002333 OXU21525.1 AAZX01000028 GAMC01009279 GAMC01009278 JAB97277.1 KB631579 ERL84447.1 APGK01058731 KB741291 ENN70573.1 NEDP02002179 OWF51662.1 CH954179 KQS62846.1 CH480816 EDW47540.1 CM000362 CM002911 EDX06741.1 KMY93165.1 AE013599 AY119635 AAF58530.1 AAM50289.1 GDRN01100882 GDRN01100881 JAI58470.1 CM000158 KRJ99466.1 ACZ94401.1 KMY93164.1 AK417692 BAN20907.1 EDV56340.2 JRES01001422 KNC23024.1 KA647797 AFP62426.1 EDW90796.2 BT072883 ACN67107.1 KK854143 PTY12052.1 CCAG010016893 GECU01030638 JAS77068.1 CH902619 KPU76085.1 JXJN01008264 CH940648 KRF79482.1 CH933808 KRG05341.1 OUUW01000001 SPP75139.1 SPP75138.1 EDW60562.2 CH963894 EDW77057.1 EDW10600.2 CH916375 EDV98188.1 EDV36209.2 CAQQ02023992 GEBQ01014912 JAT25065.1 KK852428 KDR24047.1 SPP75137.1 CP012524 ALC41850.1 GECZ01010494 GECZ01005230 JAS59275.1 JAS64539.1 CM000071 KRT01695.1 EAL24921.3 CH479181 EDW31692.1 RJVU01061435 ROJ36095.1 EDW10602.2
Proteomes
UP000005204
UP000037510
UP000218220
UP000069940
UP000249989
UP000009046
+ More
UP000069272 UP000235965 UP000008820 UP000000673 UP000192223 UP000092461 UP000075881 UP000076408 UP000075884 UP000245037 UP000075885 UP000075900 UP000076407 UP000007062 UP000075840 UP000075902 UP000075920 UP000075882 UP000030765 UP000002320 UP000075883 UP000007266 UP000075880 UP000075886 UP000075903 UP000092462 UP000215335 UP000002358 UP000030742 UP000019118 UP000242188 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000037069 UP000095301 UP000092444 UP000078200 UP000092445 UP000007801 UP000095300 UP000091820 UP000092443 UP000092460 UP000008792 UP000009192 UP000268350 UP000007798 UP000001070 UP000015102 UP000027135 UP000192221 UP000092553 UP000001819 UP000008744
UP000069272 UP000235965 UP000008820 UP000000673 UP000192223 UP000092461 UP000075881 UP000076408 UP000075884 UP000245037 UP000075885 UP000075900 UP000076407 UP000007062 UP000075840 UP000075902 UP000075920 UP000075882 UP000030765 UP000002320 UP000075883 UP000007266 UP000075880 UP000075886 UP000075903 UP000092462 UP000215335 UP000002358 UP000030742 UP000019118 UP000242188 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000037069 UP000095301 UP000092444 UP000078200 UP000092445 UP000007801 UP000095300 UP000091820 UP000092443 UP000092460 UP000008792 UP000009192 UP000268350 UP000007798 UP000001070 UP000015102 UP000027135 UP000192221 UP000092553 UP000001819 UP000008744
Interpro
IPR038765
Papain-like_cys_pep_sf
+ More
IPR003653 Peptidase_C48_C
IPR005839 Methylthiotransferase
IPR006463 MiaB_methiolase
IPR006638 Elp3/MiaB/NifB
IPR007197 rSAM
IPR038135 Methylthiotransferase_N_sf
IPR020612 Methylthiotransferase_CS
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR002792 TRAM_dom
IPR000855 Peptidase_C5
IPR003653 Peptidase_C48_C
IPR005839 Methylthiotransferase
IPR006463 MiaB_methiolase
IPR006638 Elp3/MiaB/NifB
IPR007197 rSAM
IPR038135 Methylthiotransferase_N_sf
IPR020612 Methylthiotransferase_CS
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR002792 TRAM_dom
IPR000855 Peptidase_C5
SUPFAM
SSF54001
SSF54001
Gene 3D
ProteinModelPortal
H9JHL5
A0A0L7KVL8
A0A2A4J671
S4PXU9
A0A2W1BLE0
A0A2H1VC33
+ More
A0A023EJW9 A0A182G440 A0A1Y1LFY8 E0VQE1 A0A2M4BZE8 A0A182FLI6 A0A2J7Q959 Q179X8 W5JVX8 A0A1W4X9H4 A0A1B0CV02 A0A182K2V5 A0A2M4AT16 A0A182Y6Y5 A0A182N3E4 A0A2P8ZEI2 A0A182P0U1 A0A182RLL2 A0A182X2H2 Q7QHR4 A0A182HY69 A0A182TY24 A0A182WFM6 A0A182LHC6 A0A084VR21 A0A1Q3F837 B0XF62 U5EZA9 A0A182MDY6 D6WYW7 A0A182IXQ4 A0A182QIW3 A0A182VKD7 J3JVT9 A0A1B0GQF7 A0A336M6D3 A0A034VRG2 A0A0A1XB95 A0A0K8UQ10 A0A232ET42 K7IXL0 W8BF28 U4TUK6 N6SZB8 A0A210QSH5 A0A0Q5VZI0 B4HP91 B4QCT0 Q7JUX2 A0A0P4VSF4 A0A0R1DQB9 E1JH43 A0A0J9RBU6 R4WSK8 B3NS38 A0A0L0BUX2 T1PG56 B4P5D4 C0PV27 A0A2R7VX62 A0A1B0G7S1 A0A1A9UH13 A0A1A9ZI31 A0A1B6HQW8 A0A0P9BX41 A0A1I8Q4S2 A0A1A9WPP4 A0A1A9Y0P7 A0A1B0B4D4 A0A0Q9W2Y3 A0A0Q9XAN6 A0A3B0JNJ5 A0A3B0JVP2 B4LJQ3 B4MY68 B4KT88 B4JW50 B3MCA9 T1GDP9 A0A1B6LMV8 A0A067RJR1 A0A3B0J227 A0A1W4UJH0 A0A0M4EVF7 A0A1B6GQ56 A0A0R3NR06 A0A1W4U6P0 Q292E1 B4GDN5 A0A3N0XTI3 B4KT90
A0A023EJW9 A0A182G440 A0A1Y1LFY8 E0VQE1 A0A2M4BZE8 A0A182FLI6 A0A2J7Q959 Q179X8 W5JVX8 A0A1W4X9H4 A0A1B0CV02 A0A182K2V5 A0A2M4AT16 A0A182Y6Y5 A0A182N3E4 A0A2P8ZEI2 A0A182P0U1 A0A182RLL2 A0A182X2H2 Q7QHR4 A0A182HY69 A0A182TY24 A0A182WFM6 A0A182LHC6 A0A084VR21 A0A1Q3F837 B0XF62 U5EZA9 A0A182MDY6 D6WYW7 A0A182IXQ4 A0A182QIW3 A0A182VKD7 J3JVT9 A0A1B0GQF7 A0A336M6D3 A0A034VRG2 A0A0A1XB95 A0A0K8UQ10 A0A232ET42 K7IXL0 W8BF28 U4TUK6 N6SZB8 A0A210QSH5 A0A0Q5VZI0 B4HP91 B4QCT0 Q7JUX2 A0A0P4VSF4 A0A0R1DQB9 E1JH43 A0A0J9RBU6 R4WSK8 B3NS38 A0A0L0BUX2 T1PG56 B4P5D4 C0PV27 A0A2R7VX62 A0A1B0G7S1 A0A1A9UH13 A0A1A9ZI31 A0A1B6HQW8 A0A0P9BX41 A0A1I8Q4S2 A0A1A9WPP4 A0A1A9Y0P7 A0A1B0B4D4 A0A0Q9W2Y3 A0A0Q9XAN6 A0A3B0JNJ5 A0A3B0JVP2 B4LJQ3 B4MY68 B4KT88 B4JW50 B3MCA9 T1GDP9 A0A1B6LMV8 A0A067RJR1 A0A3B0J227 A0A1W4UJH0 A0A0M4EVF7 A0A1B6GQ56 A0A0R3NR06 A0A1W4U6P0 Q292E1 B4GDN5 A0A3N0XTI3 B4KT90
PDB
2BKR
E-value=2.6381e-36,
Score=377
Ontologies
KEGG
GO
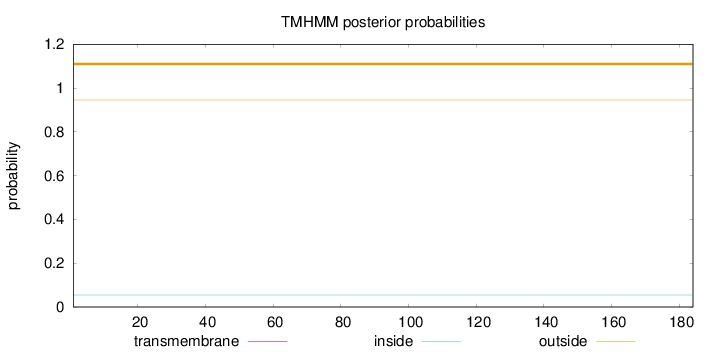
Topology
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00184
Exp number, first 60 AAs:
0.00058
Total prob of N-in:
0.05502
outside
1 - 184
Population Genetic Test Statistics
Pi
248.14477
Theta
187.804357
Tajima's D
0.662707
CLR
0.194112
CSRT
0.562721863906805
Interpretation
Uncertain
