Gene
KWMTBOMO01301
Pre Gene Modal
BGIBMGA009014
Annotation
PREDICTED:_cytochrome_b5_reductase_4_isoform_X1_[Amyelois_transitella]
Full name
Cytochrome b5 reductase 4
Alternative Name
Flavohemoprotein b5/b5R
N-terminal cytochrome b5 and cytochrome b5 oxidoreductase domain-containing protein
cb5/cb5R
N-terminal cytochrome b5 and cytochrome b5 oxidoreductase domain-containing protein
cb5/cb5R
Location in the cell
Mitochondrial Reliability : 1.917 Nuclear Reliability : 1.282
Sequence
CDS
ATGGACTGGATCAGACTGGGCAACTCCGGCAAAGATCTGACCGGCGTTGGAGGACGGATACGACCCGTCACCCACACCGAATTAGCGGCACACAACACTCAAAAAGATGCGTGGCTAGCCATCCGCGGTCGGGTTTATAACATAACCCACTACTTACCTTATCACCCTGGAGGCCCCGAAGAATTAATGCGTGGCGCAGGAATGGACGCCACCCAATTATTCGATAAAGTCCATCCTTGGGTGAACTACGACTCGCTTCTGGCCAAATGTTTAGTAGGCCCTCTGCGCTATGATCTACCCGATGCAGAAGAACTCTTCGACACCTCCTCACCATCCTCAAAATCGGACCGCCTCAGAGAACCTTCTAAAGCACAAGAATTAGTCAGAAAATCTATGGAGAATTTAGCTAATTGTATTACGCCCGTAAGAAAGAAAATATCAAATAAAAGCGAAGAAAACGCCAAAGGTAGCCCGCCGAGTAAAATAATGCAGAGTCTGATACAATCTAGCGACTTACCGGTGTCTATAAGCCGGCGAGCAGCAGCGAGTCCTGTGAAGAAAGCCGATAAAAATACAGAAACTCCCACCCCTATGCGATACGATTGGATTCAAACATCTACAAAGCTAACAATCTCCGTTTACACGGGTCCGCTGGCAAATCCTGGTGGATGTGCGAGGATTACCGACGGCTTCCTATTGATCGAAGTAGCAACGAACGGTTGGCTTAGGACACTTAAAATAACACCTGAGGCGACTTTGCAGGAACCTTTGCAGCTACGGGTGTTTGCAGAGAGTGGTAAAGTAGAGGTAATTGCACGTAAAGTGGAAAGCAGTCTCTGGAAAGGCTGCGGTGAAGTAGTCCTAGGTGTGCCAAAGAAAATCTCTTCGCCTAGAACAATGGAATGTCGTATAATGCGAGTAGATCGTGTGTCACACGACACTACATTGCTTTCGTTGACACCTAAGGCTGGTCCTGTTCTCGTGCCTCTCGGGCACCACGTACGGATACACGAGCAAATGGAAGGTTCTGAATTAGTCAGATCATATACTCCCGTAGGCGAAGGTTGGGGTGCAACTGATGACGTGCTTTCAGCTTTAAGATTAGCAGTGAAGAAATATGACTCAGGTGCAATGTCTCCGCTCTTGGCCTCTCTGAAGATTAGTGACGTGGTTACGTTATCCGGACCATATGGAAATTTTGAATTATCAAAGTTGAAAACGGTGAAGGTAATGTATCTTATAGCCGCTGGGACCGGCATTACTCCGATGCTCGGCCTCGTGAAGTTTATGCTGGCGAGATCTAACCCCAGATGTGAACGGGTGCATCTGCTTTTCTTTAACAAAACAGAACAAGACATATTATTTAATGATATCCTTGAAGATATAGCGAAGGAGGACGATAGATTTGTTGTAACAAATATTTTATCCAATGCTGGTCCATCATGGATAGGACACACAGGAAGAATATCCAAAGAACTACTTAAAAACATATTCGATAAAGATTCCATGAAATGTTATACAAAATGTACTCATTACGCTTGCATTTGTGGACCAAATGAGTTCACACATACTTCACTAGATTTGTTGAAACAAGAAGGCATGAAAGATAATTGTTTGCATGCGTTTATGGGCTAA
Protein
MDWIRLGNSGKDLTGVGGRIRPVTHTELAAHNTQKDAWLAIRGRVYNITHYLPYHPGGPEELMRGAGMDATQLFDKVHPWVNYDSLLAKCLVGPLRYDLPDAEELFDTSSPSSKSDRLREPSKAQELVRKSMENLANCITPVRKKISNKSEENAKGSPPSKIMQSLIQSSDLPVSISRRAAASPVKKADKNTETPTPMRYDWIQTSTKLTISVYTGPLANPGGCARITDGFLLIEVATNGWLRTLKITPEATLQEPLQLRVFAESGKVEVIARKVESSLWKGCGEVVLGVPKKISSPRTMECRIMRVDRVSHDTTLLSLTPKAGPVLVPLGHHVRIHEQMEGSELVRSYTPVGEGWGATDDVLSALRLAVKKYDSGAMSPLLASLKISDVVTLSGPYGNFELSKLKTVKVMYLIAAGTGITPMLGLVKFMLARSNPRCERVHLLFFNKTEQDILFNDILEDIAKEDDRFVVTNILSNAGPSWIGHTGRISKELLKNIFDKDSMKCYTKCTHYACICGPNEFTHTSLDLLKQEGMKDNCLHAFMG
Summary
Description
NADH-cytochrome b5 reductase involved in endoplasmic reticulum stress response pathway. Plays a critical role in protecting pancreatic beta-cells against oxidant stress, possibly by protecting the cell from excess buildup of reactive oxygen species (ROS).
NADH-cytochrome b5 reductase involved in endoplasmic reticulum stress response pathway.
NADH-cytochrome b5 reductase involved in endoplasmic reticulum stress response pathway.
Catalytic Activity
2 [Fe(III)-cytochrome b5] + NADH = 2 [Fe(II)-cytochrome b5] + H(+) + NAD(+)
Cofactor
FAD
Biophysicochemical Properties
25 mM for O(2)
Similarity
Belongs to the cytochrome b5 family.
Belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
Belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Endoplasmic reticulum
FAD
Flavoprotein
Heme
Iron
Metal-binding
NAD
Oxidoreductase
Polymorphism
Reference proteome
3D-structure
Feature
chain Cytochrome b5 reductase 4
splice variant In isoform 2.
sequence variant In dbSNP:rs61762820.
splice variant In isoform 2.
sequence variant In dbSNP:rs61762820.
Uniprot
H9JHL6
A0A2H1VWI4
A0A2A4J1D8
A0A2W1BMG5
A0A212ESP9
A0A194Q0R5
+ More
A0A194R9A2 A0A194RD55 A0A194PUB3 A0A1Y1KAD7 D6WYW5 A0A182WFM8 A0A182QPT9 A0A2M4BJG0 A0A182FLI8 W8BMG6 W8BHF5 A0A182Y6Y3 A0A182RLL4 A0A182N3E6 W5NIK3 W5NIK4 A0A182LHC0 A0A182LT14 A0A2M4CLH8 A0A1L8DZU9 A0A2M4CH98 K1R3R9 A0A2M4CHS2 A0A2M4CH99 A0A2M4AUB6 A0A182J5T3 A0A0K8VDC3 A0A0K8U8V0 W5UKK7 V9KPJ0 A0A2D0RK54 A0A182HY71 A0A091INN2 A0A087X649 A0A182VLE9 A0A1X7UG22 A0A3B1J0Y5 A0A182G438 Q3TDX8 A0A091W7B5 I3LV78 A0A182P0T9 A0A2K6QN53 H3ASV7 A0A3B5Q838 A0A091RA38 M4AJA2 F7DDM6 A0A3B5R1X9 U6CTX3 G7P3I7 A0A146VNU9 A0A2K5Z0F1 F7ADF0 G1PFU7 F7HJT5 S7N218 F7DDA0 A0A096NUI0 A0A2K5TRG2 A0A2K6D4F9 A0A091G9J9 A0A093TJP3 A0A091H646 A0A2U3WH08 A0A3B3QID0 A0A091IQX7 B2R7W7 A0A091QXS2 A0A2R9CF58 H2QTC4 Q7L1T6 G2HEA3 A0A3B3QJ30 A0A094KI93 A0A1L8G3B8 Q502I6 L8IHQ7 G1RQH5 W5JV99 A0A369SIU6 A0A091NE05 F1QIA4 G3QK98 A0A3B3QI07 A0A1V4JMA6 A0A0A0ALF7 B4KT92 A0A1S3RWS7 A0A1S3NHW6 A0A1E1X9K2 A0A0P4W9D4 A0A1S3RXT0
A0A194R9A2 A0A194RD55 A0A194PUB3 A0A1Y1KAD7 D6WYW5 A0A182WFM8 A0A182QPT9 A0A2M4BJG0 A0A182FLI8 W8BMG6 W8BHF5 A0A182Y6Y3 A0A182RLL4 A0A182N3E6 W5NIK3 W5NIK4 A0A182LHC0 A0A182LT14 A0A2M4CLH8 A0A1L8DZU9 A0A2M4CH98 K1R3R9 A0A2M4CHS2 A0A2M4CH99 A0A2M4AUB6 A0A182J5T3 A0A0K8VDC3 A0A0K8U8V0 W5UKK7 V9KPJ0 A0A2D0RK54 A0A182HY71 A0A091INN2 A0A087X649 A0A182VLE9 A0A1X7UG22 A0A3B1J0Y5 A0A182G438 Q3TDX8 A0A091W7B5 I3LV78 A0A182P0T9 A0A2K6QN53 H3ASV7 A0A3B5Q838 A0A091RA38 M4AJA2 F7DDM6 A0A3B5R1X9 U6CTX3 G7P3I7 A0A146VNU9 A0A2K5Z0F1 F7ADF0 G1PFU7 F7HJT5 S7N218 F7DDA0 A0A096NUI0 A0A2K5TRG2 A0A2K6D4F9 A0A091G9J9 A0A093TJP3 A0A091H646 A0A2U3WH08 A0A3B3QID0 A0A091IQX7 B2R7W7 A0A091QXS2 A0A2R9CF58 H2QTC4 Q7L1T6 G2HEA3 A0A3B3QJ30 A0A094KI93 A0A1L8G3B8 Q502I6 L8IHQ7 G1RQH5 W5JV99 A0A369SIU6 A0A091NE05 F1QIA4 G3QK98 A0A3B3QI07 A0A1V4JMA6 A0A0A0ALF7 B4KT92 A0A1S3RWS7 A0A1S3NHW6 A0A1E1X9K2 A0A0P4W9D4 A0A1S3RXT0
EC Number
1.6.2.2
Pubmed
19121390
28756777
22118469
26354079
28004739
18362917
+ More
19820115 24495485 25244985 20966253 22992520 23127152 24402279 25329095 26483478 16141072 19468303 15489334 10611283 14962668 15247412 16814408 25362486 9215903 23542700 19892987 22002653 17431167 21993624 25243066 29240929 22722832 16136131 14574404 22814378 20630863 15504981 15131110 16959974 21484476 27762356 22751099 20920257 23761445 30042472 23594743 22398555 17994087 18057021 28503490
19820115 24495485 25244985 20966253 22992520 23127152 24402279 25329095 26483478 16141072 19468303 15489334 10611283 14962668 15247412 16814408 25362486 9215903 23542700 19892987 22002653 17431167 21993624 25243066 29240929 22722832 16136131 14574404 22814378 20630863 15504981 15131110 16959974 21484476 27762356 22751099 20920257 23761445 30042472 23594743 22398555 17994087 18057021 28503490
EMBL
BABH01012801
ODYU01004705
SOQ44872.1
NWSH01004310
PCG65232.1
KZ150048
+ More
PZC74467.1 AGBW02012746 OWR44507.1 KQ459591 KPI97000.1 KQ460597 KPJ13840.1 KPJ13841.1 KPI97001.1 GEZM01087897 JAV58409.1 KQ971363 EFA07854.2 AXCN02001110 GGFJ01003950 MBW53091.1 GAMC01008384 GAMC01008382 JAB98173.1 GAMC01008383 JAB98172.1 AHAT01009243 AHAT01009244 AHAT01009245 AHAT01009246 AXCM01001435 AXCM01001436 GGFL01002012 MBW66190.1 GFDF01002165 JAV11919.1 GGFL01000539 MBW64717.1 JH817581 EKC38069.1 GGFL01000724 MBW64902.1 GGFL01000538 MBW64716.1 GGFK01011055 MBW44376.1 GDHF01015764 JAI36550.1 GDHF01029296 GDHF01027574 JAI23018.1 JAI24740.1 JT417830 AHH42516.1 JW868071 AFP00589.1 APCN01004497 KK500574 KFP09005.1 AYCK01024375 AYCK01024376 AYCK01024377 JXUM01006928 JXUM01006929 JXUM01006930 JXUM01006931 KQ560227 KXJ83737.1 AK078682 AK090159 AK157312 AK167436 AK169937 AK172252 AC156793 BC025438 BC002170 AF338818 AY321368 AY321369 KK734920 KFR11517.1 AEMK02000001 AFYH01061691 AFYH01061692 AFYH01061693 AFYH01061694 AFYH01061695 AFYH01061696 AFYH01061697 AFYH01061698 AFYH01061699 AFYH01061700 KK715023 KFQ35784.1 HAAF01001466 CCP73292.1 CM001279 EHH53210.1 GCES01067694 JAR18629.1 JSUE03031461 CM001256 EHH18505.1 AAPE02053075 AAPE02053076 AAPE02053077 AAPE02053078 AAPE02053079 GAMT01005119 GAMS01004249 GAMR01008507 GAMQ01004520 GAMP01004993 JAB06742.1 JAB18887.1 JAB25425.1 JAB37331.1 JAB47762.1 KE162930 EPQ10030.1 AHZZ02025161 AHZZ02025162 AQIA01050963 AQIA01050964 AQIA01050965 AQIA01050966 KL447974 KFO79050.1 KL442178 KFW94489.1 KL523126 KFO90292.1 KL218085 KFP02061.1 AK313146 BAG35964.1 KK684822 KFQ14144.1 AJFE02055677 AJFE02055678 AJFE02055679 AACZ04027281 GABC01004098 GABF01005424 GABE01010288 NBAG03000210 JAA07240.1 JAA16721.1 JAA34451.1 PNI88022.1 EU448291 AL034347 AL139232 BC025380 AF169803 AK305067 BAK62061.1 KL348158 KFZ57132.1 CM004475 OCT78306.1 BC095683 JH881309 ELR55069.1 ADFV01180245 ADFV01180246 ADFV01180247 ADFV01180248 ADFV01180249 ADFV01180250 ADFV01180251 ADFV01180252 ADMH02000250 ETN67243.1 NOWV01000012 RDD45933.1 KL382073 KFP87958.1 CABZ01043952 LO017673 CABD030046379 CABD030046380 CABD030046381 CABD030046382 CABD030046383 LSYS01006902 OPJ73336.1 KL872360 KGL95319.1 CH933808 EDW10604.1 KRG05344.1 GFAC01003269 JAT95919.1 GDRN01089823 JAI60595.1
PZC74467.1 AGBW02012746 OWR44507.1 KQ459591 KPI97000.1 KQ460597 KPJ13840.1 KPJ13841.1 KPI97001.1 GEZM01087897 JAV58409.1 KQ971363 EFA07854.2 AXCN02001110 GGFJ01003950 MBW53091.1 GAMC01008384 GAMC01008382 JAB98173.1 GAMC01008383 JAB98172.1 AHAT01009243 AHAT01009244 AHAT01009245 AHAT01009246 AXCM01001435 AXCM01001436 GGFL01002012 MBW66190.1 GFDF01002165 JAV11919.1 GGFL01000539 MBW64717.1 JH817581 EKC38069.1 GGFL01000724 MBW64902.1 GGFL01000538 MBW64716.1 GGFK01011055 MBW44376.1 GDHF01015764 JAI36550.1 GDHF01029296 GDHF01027574 JAI23018.1 JAI24740.1 JT417830 AHH42516.1 JW868071 AFP00589.1 APCN01004497 KK500574 KFP09005.1 AYCK01024375 AYCK01024376 AYCK01024377 JXUM01006928 JXUM01006929 JXUM01006930 JXUM01006931 KQ560227 KXJ83737.1 AK078682 AK090159 AK157312 AK167436 AK169937 AK172252 AC156793 BC025438 BC002170 AF338818 AY321368 AY321369 KK734920 KFR11517.1 AEMK02000001 AFYH01061691 AFYH01061692 AFYH01061693 AFYH01061694 AFYH01061695 AFYH01061696 AFYH01061697 AFYH01061698 AFYH01061699 AFYH01061700 KK715023 KFQ35784.1 HAAF01001466 CCP73292.1 CM001279 EHH53210.1 GCES01067694 JAR18629.1 JSUE03031461 CM001256 EHH18505.1 AAPE02053075 AAPE02053076 AAPE02053077 AAPE02053078 AAPE02053079 GAMT01005119 GAMS01004249 GAMR01008507 GAMQ01004520 GAMP01004993 JAB06742.1 JAB18887.1 JAB25425.1 JAB37331.1 JAB47762.1 KE162930 EPQ10030.1 AHZZ02025161 AHZZ02025162 AQIA01050963 AQIA01050964 AQIA01050965 AQIA01050966 KL447974 KFO79050.1 KL442178 KFW94489.1 KL523126 KFO90292.1 KL218085 KFP02061.1 AK313146 BAG35964.1 KK684822 KFQ14144.1 AJFE02055677 AJFE02055678 AJFE02055679 AACZ04027281 GABC01004098 GABF01005424 GABE01010288 NBAG03000210 JAA07240.1 JAA16721.1 JAA34451.1 PNI88022.1 EU448291 AL034347 AL139232 BC025380 AF169803 AK305067 BAK62061.1 KL348158 KFZ57132.1 CM004475 OCT78306.1 BC095683 JH881309 ELR55069.1 ADFV01180245 ADFV01180246 ADFV01180247 ADFV01180248 ADFV01180249 ADFV01180250 ADFV01180251 ADFV01180252 ADMH02000250 ETN67243.1 NOWV01000012 RDD45933.1 KL382073 KFP87958.1 CABZ01043952 LO017673 CABD030046379 CABD030046380 CABD030046381 CABD030046382 CABD030046383 LSYS01006902 OPJ73336.1 KL872360 KGL95319.1 CH933808 EDW10604.1 KRG05344.1 GFAC01003269 JAT95919.1 GDRN01089823 JAI60595.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000007266
+ More
UP000075920 UP000075886 UP000069272 UP000076408 UP000075900 UP000075884 UP000018468 UP000075882 UP000075883 UP000005408 UP000075880 UP000221080 UP000075840 UP000053119 UP000028760 UP000075903 UP000007879 UP000018467 UP000069940 UP000249989 UP000000589 UP000053605 UP000008227 UP000075885 UP000233200 UP000008672 UP000002852 UP000002281 UP000009130 UP000233140 UP000006718 UP000001074 UP000008225 UP000028761 UP000233100 UP000233120 UP000053760 UP000245340 UP000261540 UP000054308 UP000240080 UP000002277 UP000005640 UP000186698 UP000000437 UP000001073 UP000000673 UP000253843 UP000001519 UP000190648 UP000053858 UP000009192 UP000087266
UP000075920 UP000075886 UP000069272 UP000076408 UP000075900 UP000075884 UP000018468 UP000075882 UP000075883 UP000005408 UP000075880 UP000221080 UP000075840 UP000053119 UP000028760 UP000075903 UP000007879 UP000018467 UP000069940 UP000249989 UP000000589 UP000053605 UP000008227 UP000075885 UP000233200 UP000008672 UP000002852 UP000002281 UP000009130 UP000233140 UP000006718 UP000001074 UP000008225 UP000028761 UP000233100 UP000233120 UP000053760 UP000245340 UP000261540 UP000054308 UP000240080 UP000002277 UP000005640 UP000186698 UP000000437 UP000001073 UP000000673 UP000253843 UP000001519 UP000190648 UP000053858 UP000009192 UP000087266
Interpro
IPR008333
Cbr1-like_FAD-bd_dom
+ More
IPR039261 FNR_nucleotide-bd
IPR018506 Cyt_B5_heme-BS
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR017927 FAD-bd_FR_type
IPR001433 OxRdtase_FAD/NAD-bd
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR037908 p23_NCB5OR
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR039261 FNR_nucleotide-bd
IPR018506 Cyt_B5_heme-BS
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR017927 FAD-bd_FR_type
IPR001433 OxRdtase_FAD/NAD-bd
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR037908 p23_NCB5OR
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
Gene 3D
ProteinModelPortal
H9JHL6
A0A2H1VWI4
A0A2A4J1D8
A0A2W1BMG5
A0A212ESP9
A0A194Q0R5
+ More
A0A194R9A2 A0A194RD55 A0A194PUB3 A0A1Y1KAD7 D6WYW5 A0A182WFM8 A0A182QPT9 A0A2M4BJG0 A0A182FLI8 W8BMG6 W8BHF5 A0A182Y6Y3 A0A182RLL4 A0A182N3E6 W5NIK3 W5NIK4 A0A182LHC0 A0A182LT14 A0A2M4CLH8 A0A1L8DZU9 A0A2M4CH98 K1R3R9 A0A2M4CHS2 A0A2M4CH99 A0A2M4AUB6 A0A182J5T3 A0A0K8VDC3 A0A0K8U8V0 W5UKK7 V9KPJ0 A0A2D0RK54 A0A182HY71 A0A091INN2 A0A087X649 A0A182VLE9 A0A1X7UG22 A0A3B1J0Y5 A0A182G438 Q3TDX8 A0A091W7B5 I3LV78 A0A182P0T9 A0A2K6QN53 H3ASV7 A0A3B5Q838 A0A091RA38 M4AJA2 F7DDM6 A0A3B5R1X9 U6CTX3 G7P3I7 A0A146VNU9 A0A2K5Z0F1 F7ADF0 G1PFU7 F7HJT5 S7N218 F7DDA0 A0A096NUI0 A0A2K5TRG2 A0A2K6D4F9 A0A091G9J9 A0A093TJP3 A0A091H646 A0A2U3WH08 A0A3B3QID0 A0A091IQX7 B2R7W7 A0A091QXS2 A0A2R9CF58 H2QTC4 Q7L1T6 G2HEA3 A0A3B3QJ30 A0A094KI93 A0A1L8G3B8 Q502I6 L8IHQ7 G1RQH5 W5JV99 A0A369SIU6 A0A091NE05 F1QIA4 G3QK98 A0A3B3QI07 A0A1V4JMA6 A0A0A0ALF7 B4KT92 A0A1S3RWS7 A0A1S3NHW6 A0A1E1X9K2 A0A0P4W9D4 A0A1S3RXT0
A0A194R9A2 A0A194RD55 A0A194PUB3 A0A1Y1KAD7 D6WYW5 A0A182WFM8 A0A182QPT9 A0A2M4BJG0 A0A182FLI8 W8BMG6 W8BHF5 A0A182Y6Y3 A0A182RLL4 A0A182N3E6 W5NIK3 W5NIK4 A0A182LHC0 A0A182LT14 A0A2M4CLH8 A0A1L8DZU9 A0A2M4CH98 K1R3R9 A0A2M4CHS2 A0A2M4CH99 A0A2M4AUB6 A0A182J5T3 A0A0K8VDC3 A0A0K8U8V0 W5UKK7 V9KPJ0 A0A2D0RK54 A0A182HY71 A0A091INN2 A0A087X649 A0A182VLE9 A0A1X7UG22 A0A3B1J0Y5 A0A182G438 Q3TDX8 A0A091W7B5 I3LV78 A0A182P0T9 A0A2K6QN53 H3ASV7 A0A3B5Q838 A0A091RA38 M4AJA2 F7DDM6 A0A3B5R1X9 U6CTX3 G7P3I7 A0A146VNU9 A0A2K5Z0F1 F7ADF0 G1PFU7 F7HJT5 S7N218 F7DDA0 A0A096NUI0 A0A2K5TRG2 A0A2K6D4F9 A0A091G9J9 A0A093TJP3 A0A091H646 A0A2U3WH08 A0A3B3QID0 A0A091IQX7 B2R7W7 A0A091QXS2 A0A2R9CF58 H2QTC4 Q7L1T6 G2HEA3 A0A3B3QJ30 A0A094KI93 A0A1L8G3B8 Q502I6 L8IHQ7 G1RQH5 W5JV99 A0A369SIU6 A0A091NE05 F1QIA4 G3QK98 A0A3B3QI07 A0A1V4JMA6 A0A0A0ALF7 B4KT92 A0A1S3RWS7 A0A1S3NHW6 A0A1E1X9K2 A0A0P4W9D4 A0A1S3RXT0
PDB
3LF5
E-value=2.68588e-23,
Score=270
Ontologies
GO
GO:0016491
GO:0046872
GO:0020037
GO:0016021
GO:0004128
GO:0055114
GO:0005783
GO:0006801
GO:0006091
GO:0016653
GO:0030073
GO:0016174
GO:0042168
GO:0005737
GO:0006739
GO:0003958
GO:0048471
GO:0048468
GO:0050660
GO:0046677
GO:0042593
GO:0015701
GO:0005789
GO:0003032
GO:0000287
GO:0015934
GO:0005576
GO:0016042
GO:0005634
GO:0006334
GO:0003779
Topology
Subcellular location
Endoplasmic reticulum
Soluble protein. With evidence from 1 publications.
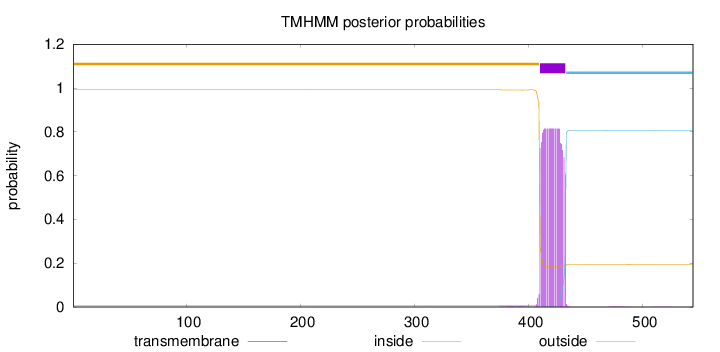
Length:
544
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.21198
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.00758
outside
1 - 409
TMhelix
410 - 432
inside
433 - 544
Population Genetic Test Statistics
Pi
246.641816
Theta
175.798129
Tajima's D
1.414148
CLR
0.417238
CSRT
0.770461476926154
Interpretation
Uncertain
