Pre Gene Modal
BGIBMGA008874
Annotation
PREDICTED:_FAS-associated_factor_2_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.351
Sequence
CDS
ATGGATCTTGAAGACAATGCTTTGGGTTTGACCCAGGATCAGACCGATAAAATTTTACAATTTCAAGACTTAACTGGCATTGAAGACATGTCAATATGTAGAGATGTTTTGCAGAGACACCAATGGGATCTAGAGGTAGCAATTCAAGAACAATTGAATATTAGAGAAGGGCGCCCCTCTGTGTTTGCCACAGAAGCTCGGGCACCACCTGTTGTACATGACCACATTGCACAGCAAGTTTTTACTGAAGAACCACCAGAGGGACCCGGAGGTGTCAGGGGCCTAATACGTTATGTTGTTAATCTAGTTGTCTCAATGTGCTATAGTACAATTTCATCTGTATTAAACTTGCTTTTAAGTTTTGTACGTAATGATGATAGAAGACTGGCAACTGATCCATTAGCAGATGTAATGAATTTCATCAGTAGCTACCAAACAAAATTCAATGCACACCCAGTCTTCTATCAGGGAACTTATGCTCAAGCTCTAAACGATGCTAAGAACGAGCTCAGATTCCTAATTGTTTACTTACATTCAGATTCTGCACCAGAAACACAGAATTTCTGCAGAACAACATTAGCAGATCCCGAAGTAGTCCAATATATAAATACGCATGCATTGTTCTGGGGTTGCTCAGTCGAGAGTGGGGAAGGTTGGCGCGTCGCTCAGTCGGTGGGAGGTCGGCGATATCCTCTTTTGTGCGTTGTCTGCGTGCGGGAACATCGCATGACTGTAGTCGCTCGAAGTGAAGGGGCCTGTTCCGCCGAAATACTTATGCAGCGTCTTCGTCAAGTTGTCAGTGAAAACGAGGGTCACTTGGCTGCCGCTCGAGCGGATAGAGTGGAACGCGAAGTGACAGCGCGGCTGAGGGCTGCCCAGGACGAAGCATACGCCGAATCCCTCGCGGCCGACCAGGAGAAGGAGAGACGAAAAGCGGCCGCCCGTGAATTAGAACAGTTACAGCAACAACAGTACTTAGAGGAATTGGAGCAAGAGGAACGCCGGAAACAAGAGGTGGTGGAGGCTCGTAAAGCGATGGCGTCTCGTCTGCCGTTGGAGCCGTCGGGCGGCGCTGACACCGTGGTGCTTCTGATCCGTCTGCCAGACGGCGAACGTCTCACTCGCAGATTTTCACATTTCCACACCACTCAGGATCTATACGACTTTGTGTTCAGCCATCCGCAGTCTCCGGAAGAATTCGAGATAACGACAAACTTTCCCAAACGCGTGTTAGCCCGAGGAGCGTCCAACTTGTTGGACGTGGGACTGAAGGATCGAGACGTGTTGTTCGTTAACGATATCAACGCTTGA
Protein
MDLEDNALGLTQDQTDKILQFQDLTGIEDMSICRDVLQRHQWDLEVAIQEQLNIREGRPSVFATEARAPPVVHDHIAQQVFTEEPPEGPGGVRGLIRYVVNLVVSMCYSTISSVLNLLLSFVRNDDRRLATDPLADVMNFISSYQTKFNAHPVFYQGTYAQALNDAKNELRFLIVYLHSDSAPETQNFCRTTLADPEVVQYINTHALFWGCSVESGEGWRVAQSVGGRRYPLLCVVCVREHRMTVVARSEGACSAEILMQRLRQVVSENEGHLAAARADRVEREVTARLRAAQDEAYAESLAADQEKERRKAAARELEQLQQQQYLEELEQEERRKQEVVEARKAMASRLPLEPSGGADTVVLLIRLPDGERLTRRFSHFHTTQDLYDFVFSHPQSPEEFEITTNFPKRVLARGASNLLDVGLKDRDVLFVNDINA
Summary
Uniprot
H9JH77
A0A2H1VVK1
A0A194R871
A0A2W1BHY7
A0A212F4G4
A0A2A4J1H4
+ More
A0A1Q3F6M6 A0A1Q3F6M0 B0XAP2 U5EWD1 Q17DA0 A0A3B0K8X8 A0A0K8TN65 A0A0M4ERX1 B3MM71 A0A023ETD9 Q29LE0 A0A1L8DM56 A0A1W4VM42 A0A182GTC5 A0A182RJ99 E0VQE6 B4I5K1 A0A182M8B0 B4P9X3 A0A2M4AK37 B3NLA5 A0A182W7G8 A0A2M3Z9L3 A0A2M4BNG2 W5JDM5 B4MYS8 A0A182XZV5 A0A182JEN8 A0A182PD12 Q9VJ58 A0A182S9A8 A0A182NTK4 B4LTS3 A0A084W5Y0 O77047 B4Q8X6 A0A1B6C6X3 A0A182JXT4 B4JAN7 W8APP1 A0A1A9UNR8 D3TN73 A0A0L0BQZ7 A0A1Y1LTJ6 A0A1A9W0Q3 A0A1B0C1N7 A0A0A1WWA7 A0A1L8DM72 A0A0K8URV1 A0A1A9YIS5 A0A067RA97 A0A034VZP4 A0A182F1I9 A0A1I8MWG9 A0A182U3I4 T1PC86 T1PL87 A0A336M8X6 B4KGA1 A0A1B0A7N0 V5GZD6 A0A182QSJ4 A0A1I8P390 A0A224XCV5 A0A182UM10 A0A182WT35 A0A182L1Y5 Q7Q3P6 A0A182HV32 A0A1B6EXC1 A0A1B0GB94 A0A1B6LIR6 A0A1B6M7B8 A0A069DTI1 A0A1W4WYR3 A0A0V0G8G6 A0A023F8M5 A0A1B0CVU1 A0A2J7QQC2 A0A1J1HZ92 E2BW74 U4U2Z3 A0A0N7Z985 R4FLP8 A0A195DSM1 A0A2M4AL85 D2CG01 A0A195ATY1 A0A026WQN2 A0A0A9XUE8 A0A158NPV1
A0A1Q3F6M6 A0A1Q3F6M0 B0XAP2 U5EWD1 Q17DA0 A0A3B0K8X8 A0A0K8TN65 A0A0M4ERX1 B3MM71 A0A023ETD9 Q29LE0 A0A1L8DM56 A0A1W4VM42 A0A182GTC5 A0A182RJ99 E0VQE6 B4I5K1 A0A182M8B0 B4P9X3 A0A2M4AK37 B3NLA5 A0A182W7G8 A0A2M3Z9L3 A0A2M4BNG2 W5JDM5 B4MYS8 A0A182XZV5 A0A182JEN8 A0A182PD12 Q9VJ58 A0A182S9A8 A0A182NTK4 B4LTS3 A0A084W5Y0 O77047 B4Q8X6 A0A1B6C6X3 A0A182JXT4 B4JAN7 W8APP1 A0A1A9UNR8 D3TN73 A0A0L0BQZ7 A0A1Y1LTJ6 A0A1A9W0Q3 A0A1B0C1N7 A0A0A1WWA7 A0A1L8DM72 A0A0K8URV1 A0A1A9YIS5 A0A067RA97 A0A034VZP4 A0A182F1I9 A0A1I8MWG9 A0A182U3I4 T1PC86 T1PL87 A0A336M8X6 B4KGA1 A0A1B0A7N0 V5GZD6 A0A182QSJ4 A0A1I8P390 A0A224XCV5 A0A182UM10 A0A182WT35 A0A182L1Y5 Q7Q3P6 A0A182HV32 A0A1B6EXC1 A0A1B0GB94 A0A1B6LIR6 A0A1B6M7B8 A0A069DTI1 A0A1W4WYR3 A0A0V0G8G6 A0A023F8M5 A0A1B0CVU1 A0A2J7QQC2 A0A1J1HZ92 E2BW74 U4U2Z3 A0A0N7Z985 R4FLP8 A0A195DSM1 A0A2M4AL85 D2CG01 A0A195ATY1 A0A026WQN2 A0A0A9XUE8 A0A158NPV1
Pubmed
19121390
26354079
28756777
22118469
17510324
26369729
+ More
17994087 24945155 26483478 15632085 23185243 20566863 17550304 20920257 23761445 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 22936249 24495485 20353571 26108605 28004739 25830018 24845553 25348373 25315136 20966253 12364791 14747013 17210077 26334808 25474469 20798317 23537049 27129103 18362917 19820115 24508170 30249741 25401762 26823975 21347285
17994087 24945155 26483478 15632085 23185243 20566863 17550304 20920257 23761445 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 22936249 24495485 20353571 26108605 28004739 25830018 24845553 25348373 25315136 20966253 12364791 14747013 17210077 26334808 25474469 20798317 23537049 27129103 18362917 19820115 24508170 30249741 25401762 26823975 21347285
EMBL
BABH01012801
BABH01012802
BABH01012803
ODYU01004705
SOQ44871.1
KQ460597
+ More
KPJ13842.1 KZ150048 PZC74468.1 AGBW02010367 OWR48635.1 NWSH01004310 PCG65233.1 GFDL01011824 JAV23221.1 GFDL01011830 JAV23215.1 DS232592 EDS43831.1 GANO01001470 JAB58401.1 CH477298 EAT44329.1 OUUW01000004 SPP79978.1 GDAI01001804 JAI15799.1 CP012523 ALC39987.1 CH902620 EDV30886.1 JXUM01000242 JXUM01000243 JXUM01000244 JXUM01000245 GAPW01001487 JAC12111.1 CH379060 EAL34105.1 KRT04474.1 GFDF01006531 JAV07553.1 JXUM01017940 JXUM01017941 DS235418 EEB15602.1 CH480822 EDW55657.1 AXCM01003126 CM000158 EDW90314.1 GGFK01007820 MBW41141.1 CH954179 EDV54755.1 GGFM01004476 MBW25227.1 GGFJ01005362 MBW54503.1 ADMH02001688 ETN61433.1 CH963913 EDW77267.1 AE014134 AY051515 AAF53697.1 AAK92939.1 AHN54571.1 CH940649 EDW63974.1 ATLV01020706 KE525305 KFB45624.1 AB013610 BAA33466.1 CM000361 CM002910 EDX05408.1 KMY90866.1 GEDC01028144 GEDC01016573 JAS09154.1 JAS20725.1 CH916368 EDW03845.1 GAMC01018558 JAB87997.1 EZ422875 ADD19151.1 JRES01001507 KNC22403.1 GEZM01046895 JAV76972.1 JXJN01024139 GBXI01010978 JAD03314.1 GFDF01006532 JAV07552.1 GDHF01023234 GDHF01010745 JAI29080.1 JAI41569.1 KK852639 KDR19659.1 GAKP01011919 JAC47033.1 KA645548 AFP60177.1 KA649527 AFP64156.1 UFQT01000502 SSX24817.1 CH933807 EDW11088.1 GALX01002608 JAB65858.1 AXCN02000795 GFTR01006199 JAW10227.1 AAAB01008964 EAA12408.2 APCN01002491 GECZ01027212 JAS42557.1 CCAG010011564 GEBQ01016427 JAT23550.1 GEBQ01008147 JAT31830.1 GBGD01001526 JAC87363.1 GECL01001983 JAP04141.1 GBBI01001000 JAC17712.1 AJWK01031243 NEVH01012087 PNF30776.1 CVRI01000037 CRK93275.1 GL451091 EFN80106.1 KB631948 ERL87412.1 GDKW01001251 JAI55344.1 GAHY01001293 JAA76217.1 KQ980487 KYN15915.1 GGFK01008228 MBW41549.1 DS497677 EFA11847.1 KQ976745 KYM75480.1 KK107148 QOIP01000009 EZA57414.1 RLU18772.1 GBHO01022806 GBHO01022794 GBRD01017890 GDHC01004407 JAG20798.1 JAG20810.1 JAG47937.1 JAQ14222.1 ADTU01022757
KPJ13842.1 KZ150048 PZC74468.1 AGBW02010367 OWR48635.1 NWSH01004310 PCG65233.1 GFDL01011824 JAV23221.1 GFDL01011830 JAV23215.1 DS232592 EDS43831.1 GANO01001470 JAB58401.1 CH477298 EAT44329.1 OUUW01000004 SPP79978.1 GDAI01001804 JAI15799.1 CP012523 ALC39987.1 CH902620 EDV30886.1 JXUM01000242 JXUM01000243 JXUM01000244 JXUM01000245 GAPW01001487 JAC12111.1 CH379060 EAL34105.1 KRT04474.1 GFDF01006531 JAV07553.1 JXUM01017940 JXUM01017941 DS235418 EEB15602.1 CH480822 EDW55657.1 AXCM01003126 CM000158 EDW90314.1 GGFK01007820 MBW41141.1 CH954179 EDV54755.1 GGFM01004476 MBW25227.1 GGFJ01005362 MBW54503.1 ADMH02001688 ETN61433.1 CH963913 EDW77267.1 AE014134 AY051515 AAF53697.1 AAK92939.1 AHN54571.1 CH940649 EDW63974.1 ATLV01020706 KE525305 KFB45624.1 AB013610 BAA33466.1 CM000361 CM002910 EDX05408.1 KMY90866.1 GEDC01028144 GEDC01016573 JAS09154.1 JAS20725.1 CH916368 EDW03845.1 GAMC01018558 JAB87997.1 EZ422875 ADD19151.1 JRES01001507 KNC22403.1 GEZM01046895 JAV76972.1 JXJN01024139 GBXI01010978 JAD03314.1 GFDF01006532 JAV07552.1 GDHF01023234 GDHF01010745 JAI29080.1 JAI41569.1 KK852639 KDR19659.1 GAKP01011919 JAC47033.1 KA645548 AFP60177.1 KA649527 AFP64156.1 UFQT01000502 SSX24817.1 CH933807 EDW11088.1 GALX01002608 JAB65858.1 AXCN02000795 GFTR01006199 JAW10227.1 AAAB01008964 EAA12408.2 APCN01002491 GECZ01027212 JAS42557.1 CCAG010011564 GEBQ01016427 JAT23550.1 GEBQ01008147 JAT31830.1 GBGD01001526 JAC87363.1 GECL01001983 JAP04141.1 GBBI01001000 JAC17712.1 AJWK01031243 NEVH01012087 PNF30776.1 CVRI01000037 CRK93275.1 GL451091 EFN80106.1 KB631948 ERL87412.1 GDKW01001251 JAI55344.1 GAHY01001293 JAA76217.1 KQ980487 KYN15915.1 GGFK01008228 MBW41549.1 DS497677 EFA11847.1 KQ976745 KYM75480.1 KK107148 QOIP01000009 EZA57414.1 RLU18772.1 GBHO01022806 GBHO01022794 GBRD01017890 GDHC01004407 JAG20798.1 JAG20810.1 JAG47937.1 JAQ14222.1 ADTU01022757
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000002320
UP000008820
+ More
UP000268350 UP000092553 UP000007801 UP000069940 UP000001819 UP000192221 UP000075900 UP000009046 UP000001292 UP000075883 UP000002282 UP000008711 UP000075920 UP000000673 UP000007798 UP000076408 UP000075880 UP000075885 UP000000803 UP000075901 UP000075884 UP000008792 UP000030765 UP000000304 UP000075881 UP000001070 UP000078200 UP000037069 UP000091820 UP000092460 UP000092443 UP000027135 UP000069272 UP000095301 UP000075902 UP000009192 UP000092445 UP000075886 UP000095300 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000092444 UP000192223 UP000092461 UP000235965 UP000183832 UP000008237 UP000030742 UP000078492 UP000007266 UP000078540 UP000053097 UP000279307 UP000005205
UP000268350 UP000092553 UP000007801 UP000069940 UP000001819 UP000192221 UP000075900 UP000009046 UP000001292 UP000075883 UP000002282 UP000008711 UP000075920 UP000000673 UP000007798 UP000076408 UP000075880 UP000075885 UP000000803 UP000075901 UP000075884 UP000008792 UP000030765 UP000000304 UP000075881 UP000001070 UP000078200 UP000037069 UP000091820 UP000092460 UP000092443 UP000027135 UP000069272 UP000095301 UP000075902 UP000009192 UP000092445 UP000075886 UP000095300 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000092444 UP000192223 UP000092461 UP000235965 UP000183832 UP000008237 UP000030742 UP000078492 UP000007266 UP000078540 UP000053097 UP000279307 UP000005205
Interpro
IPR001012
UBX_dom
+ More
IPR036249 Thioredoxin-like_sf
IPR006577 UAS
IPR029071 Ubiquitin-like_domsf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR019874 Release_fac_Glu-N5_MeTfrase
IPR040758 PrmC_N
IPR007848 Small_mtfrase_dom
IPR029063 SAM-dependent_MTases
IPR002052 DNA_methylase_N6_adenine_CS
IPR004556 HemK-like
IPR009060 UBA-like_sf
IPR036249 Thioredoxin-like_sf
IPR006577 UAS
IPR029071 Ubiquitin-like_domsf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR019874 Release_fac_Glu-N5_MeTfrase
IPR040758 PrmC_N
IPR007848 Small_mtfrase_dom
IPR029063 SAM-dependent_MTases
IPR002052 DNA_methylase_N6_adenine_CS
IPR004556 HemK-like
IPR009060 UBA-like_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JH77
A0A2H1VVK1
A0A194R871
A0A2W1BHY7
A0A212F4G4
A0A2A4J1H4
+ More
A0A1Q3F6M6 A0A1Q3F6M0 B0XAP2 U5EWD1 Q17DA0 A0A3B0K8X8 A0A0K8TN65 A0A0M4ERX1 B3MM71 A0A023ETD9 Q29LE0 A0A1L8DM56 A0A1W4VM42 A0A182GTC5 A0A182RJ99 E0VQE6 B4I5K1 A0A182M8B0 B4P9X3 A0A2M4AK37 B3NLA5 A0A182W7G8 A0A2M3Z9L3 A0A2M4BNG2 W5JDM5 B4MYS8 A0A182XZV5 A0A182JEN8 A0A182PD12 Q9VJ58 A0A182S9A8 A0A182NTK4 B4LTS3 A0A084W5Y0 O77047 B4Q8X6 A0A1B6C6X3 A0A182JXT4 B4JAN7 W8APP1 A0A1A9UNR8 D3TN73 A0A0L0BQZ7 A0A1Y1LTJ6 A0A1A9W0Q3 A0A1B0C1N7 A0A0A1WWA7 A0A1L8DM72 A0A0K8URV1 A0A1A9YIS5 A0A067RA97 A0A034VZP4 A0A182F1I9 A0A1I8MWG9 A0A182U3I4 T1PC86 T1PL87 A0A336M8X6 B4KGA1 A0A1B0A7N0 V5GZD6 A0A182QSJ4 A0A1I8P390 A0A224XCV5 A0A182UM10 A0A182WT35 A0A182L1Y5 Q7Q3P6 A0A182HV32 A0A1B6EXC1 A0A1B0GB94 A0A1B6LIR6 A0A1B6M7B8 A0A069DTI1 A0A1W4WYR3 A0A0V0G8G6 A0A023F8M5 A0A1B0CVU1 A0A2J7QQC2 A0A1J1HZ92 E2BW74 U4U2Z3 A0A0N7Z985 R4FLP8 A0A195DSM1 A0A2M4AL85 D2CG01 A0A195ATY1 A0A026WQN2 A0A0A9XUE8 A0A158NPV1
A0A1Q3F6M6 A0A1Q3F6M0 B0XAP2 U5EWD1 Q17DA0 A0A3B0K8X8 A0A0K8TN65 A0A0M4ERX1 B3MM71 A0A023ETD9 Q29LE0 A0A1L8DM56 A0A1W4VM42 A0A182GTC5 A0A182RJ99 E0VQE6 B4I5K1 A0A182M8B0 B4P9X3 A0A2M4AK37 B3NLA5 A0A182W7G8 A0A2M3Z9L3 A0A2M4BNG2 W5JDM5 B4MYS8 A0A182XZV5 A0A182JEN8 A0A182PD12 Q9VJ58 A0A182S9A8 A0A182NTK4 B4LTS3 A0A084W5Y0 O77047 B4Q8X6 A0A1B6C6X3 A0A182JXT4 B4JAN7 W8APP1 A0A1A9UNR8 D3TN73 A0A0L0BQZ7 A0A1Y1LTJ6 A0A1A9W0Q3 A0A1B0C1N7 A0A0A1WWA7 A0A1L8DM72 A0A0K8URV1 A0A1A9YIS5 A0A067RA97 A0A034VZP4 A0A182F1I9 A0A1I8MWG9 A0A182U3I4 T1PC86 T1PL87 A0A336M8X6 B4KGA1 A0A1B0A7N0 V5GZD6 A0A182QSJ4 A0A1I8P390 A0A224XCV5 A0A182UM10 A0A182WT35 A0A182L1Y5 Q7Q3P6 A0A182HV32 A0A1B6EXC1 A0A1B0GB94 A0A1B6LIR6 A0A1B6M7B8 A0A069DTI1 A0A1W4WYR3 A0A0V0G8G6 A0A023F8M5 A0A1B0CVU1 A0A2J7QQC2 A0A1J1HZ92 E2BW74 U4U2Z3 A0A0N7Z985 R4FLP8 A0A195DSM1 A0A2M4AL85 D2CG01 A0A195ATY1 A0A026WQN2 A0A0A9XUE8 A0A158NPV1
PDB
2DAM
E-value=2.95162e-11,
Score=165
Ontologies
GO
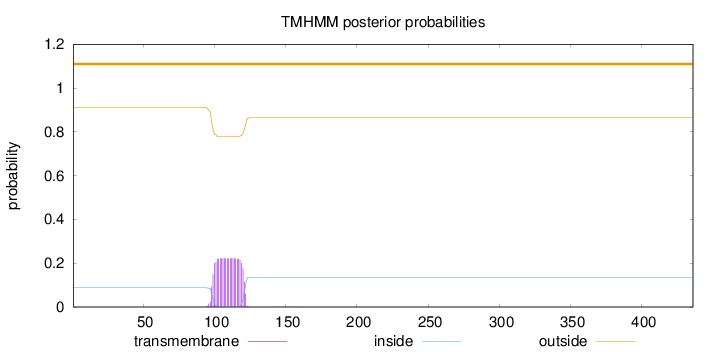
Topology
Length:
436
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.0242
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08804
outside
1 - 436
Population Genetic Test Statistics
Pi
235.187585
Theta
163.078294
Tajima's D
1.527726
CLR
0.06268
CSRT
0.793760311984401
Interpretation
Uncertain
