Gene
KWMTBOMO01299
Annotation
Retrovirus-related_Pol_polyprotein_from_transposon_TNT_1-94_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.718
Sequence
CDS
ATGTTCCTCACAGATTGTGATCTATGGGACTGTGTAAGTACAGACACACTTGATACTCCTGAATTAAAAAAGAGAGATAGTAAAGCAAGAGCAAAAATATGTCTACTCTTGGAAGAAAACATTTTCCCAATTGTTACCGGTGCCGTAACGGCAAAACAAACATGGGATGCGTTAGAAAAGGCCTACGCCGATGGCGGACTGTTAAGACGACTCACTTTATTAAGAAAATTATTTAATTTGAAATTATCATGTTTCGTAAATATCGAAGCATACATCAATGAAGTGCAGGCGACAAGTCAGGCTTTACAGCGAATCAAATCAGGACTGGATGATGAATTCATTGGGATCATAATGTTAGCTGGGTTGACCGAGGAATACAATCCTTTGGTGATGGCGGTGGAACATTCTGAAAGGAAATTAACTTCAGAAGCGGTCATATCTGCTCTAATGAAGGAAGGACAGCGCAAGCAGCCGGCAGTTGAAATGGATTCTGTGTTAGTGTCTAATAACGAAAAACCTAGTAGTAGTACGAAAAAGGCAATAATATGTCATTTTTGTAAAAAGAAAGGTCATTTTAAATGGCAATGTAAATTTTATAAAGATAGGGATAAGAAAGGTAATGCAAATGTATGCGTAGCTCAAAAGGAGACTAATGATTTTATTACCTTACTCTCTAATATGGGAAAGTCGAGAAATTGGATTATAGATTCTGGGGCAACATCTCATATGACATCATCAGATAATGTTTTAGTAAATGCTAAAGGTGGTTACCGGAAGTATATAACTACAGCTAATAATGAGAAAATATTATCTAAGGTAAAGGGTGATGTATTGATGCCTAATTATAAGTTAAAGTTTGAGAATGTTTTGTACATACCAGATTTGATAACCAATTTACTTTCTGTTAGTAGTATTTGTAAGCATGATTGTACAGTTGTTTTTGATAAACATTCTTGTAAAGTTTACAAATCTGCAGATGTATCCTTAGTGGGAAAAACTATTTTAGAAGCACCTTTAAAAGATGGGCTATATGTAGTAGACCGAGATGACAAGGCATTGGTTACTAAAGACATTAATTCGCCTGTTAACGATCAATATAAGTTATGGCATAGGCGTTTAGGTCATTTATCACCAGCTCTAATGAATAAATTAAGAGGGTTGGTTGATGGCTTTACCTTGAATGAATTTAGTAAGGAACCATGCGTCAGTTGTTGTAAAGGTAAACAGTGCGTAAAAAAGTTCCCAAAAAGAAAGAATAAAAGAGCTAAGGATATATTGAGGATATATATGGAGGAAGAGACATGGGGTGGTGCTCGATATTTTCTTATCTTTGTTGACGATTTTAGTCGGAAAGTTATGGGATATATGTTGAAATCCAAAACAGAAGTATTTGCTAAATTTAAGGAATTCAAGGTGTTTGTGGAGAATCAAACTAATAAAAAGATCAAAAAATTGCGTAGCGATAATGGTACTGAATATATAAACGATGAATTTCAAAGCTTTCTACGAGCTAATGGTATCCATCATCAGACTTCAGTGGCACACACTCCACAACAAAATGGAGTGGCAGAGCGTACAATTCGTACTGTAATTGAAAAGGCACGTTATCCTAAATCAGTTGAGGACGCATTGGAACGTTCGGATCGAGAGTTATGGCACACAGCCATGTTAGATGGATACAATTCGCTGATGCAAAACAAAAACAGTGAACAGAATTAA
Protein
MFLTDCDLWDCVSTDTLDTPELKKRDSKARAKICLLLEENIFPIVTGAVTAKQTWDALEKAYADGGLLRRLTLLRKLFNLKLSCFVNIEAYINEVQATSQALQRIKSGLDDEFIGIIMLAGLTEEYNPLVMAVEHSERKLTSEAVISALMKEGQRKQPAVEMDSVLVSNNEKPSSSTKKAIICHFCKKKGHFKWQCKFYKDRDKKGNANVCVAQKETNDFITLLSNMGKSRNWIIDSGATSHMTSSDNVLVNAKGGYRKYITTANNEKILSKVKGDVLMPNYKLKFENVLYIPDLITNLLSVSSICKHDCTVVFDKHSCKVYKSADVSLVGKTILEAPLKDGLYVVDRDDKALVTKDINSPVNDQYKLWHRRLGHLSPALMNKLRGLVDGFTLNEFSKEPCVSCCKGKQCVKKFPKRKNKRAKDILRIYMEEETWGGARYFLIFVDDFSRKVMGYMLKSKTEVFAKFKEFKVFVENQTNKKIKKLRSDNGTEYINDEFQSFLRANGIHHQTSVAHTPQQNGVAERTIRTVIEKARYPKSVEDALERSDRELWHTAMLDGYNSLMQNKNSEQN
Summary
Uniprot
A0A0N1ICY1
A0A0A9YFM0
A0A0J7NHJ8
A0A0V0T2I4
A0A2A4J959
A0A1Y1LWI3
+ More
A0A0V1K1Y9 A0A0V1F2E6 E5SB91 A0A0V1APS7 A0A2A4JRI1 A0A0V1FKH2 A0A0V0T3I0 A0A034VP70 A0A0V0TUP0 A0A1Y1N6L8 A0A0V0WR33 A0A0A1WXQ3 A0A1Y1N668 A0A1Y1N932 A0A0N1IFR1 W4VRQ4 A0A194RJV9 A0A085MVP5 A0A0V0WQU6 A0A085N572 A0A194RU38 A0A212F7V4 A0A0A9XEY5 A0A1W7R657 W4VRP9 W4VRP2 A0A1W7R6H6 A0A224XA26 A0A0V0Z6Q0 A0A1W7R670 A0A2J7Q1Z2 A0A1W7R663 A0A2J7RKJ6 A0A2J7R668 A0A0A9Z683 A0A146LFV8 A0A2J7R8S8 A0A0V0RDY1 V5GAG2 A0A1B6E9D0 A0A0J7K0M9 A0A182GZK6 A0A1W7R6J6 A0A1W7R6K6 A0A0V0YAP7 A0A1Y1N3X2 A0A0V0YJB3 A0A0A9WQD4 A0A0V1HT08 A0A0V0YNF4 A0A0J7K5G8 K7J5Z3 A0A1W7R6D6 A0A0A9XVJ9 A0A0A9WPH5 A0A0S7KIW8 K7JBY9 A0A0K8U6S2 A0A0V1FXR9 A0A0A9WRE3 A0A0K8U956 A0A0A9XCL0 W8BYI7 A0A0A9WG89 A0A0A9WJ72 W8BFA2 A0A0J7K7B7 W8B528 W8AUM1 A0A0A1XSW5 A0A0A1WGJ2 W8ANX7 A0A0P6IXK4 K7JXM1 A0A034W7T5 A0A0A1X768 A0A2R6NZJ8 A0A0A1XLZ7 A0A182GNT8 A0A0J7K4F4 T1PKV2 W8BEX4 A0A2A4IIH8 A0A0V0V098 A0A251TN47 A0A0V1GS48 A0A131XNF9 B6V6Z8
A0A0V1K1Y9 A0A0V1F2E6 E5SB91 A0A0V1APS7 A0A2A4JRI1 A0A0V1FKH2 A0A0V0T3I0 A0A034VP70 A0A0V0TUP0 A0A1Y1N6L8 A0A0V0WR33 A0A0A1WXQ3 A0A1Y1N668 A0A1Y1N932 A0A0N1IFR1 W4VRQ4 A0A194RJV9 A0A085MVP5 A0A0V0WQU6 A0A085N572 A0A194RU38 A0A212F7V4 A0A0A9XEY5 A0A1W7R657 W4VRP9 W4VRP2 A0A1W7R6H6 A0A224XA26 A0A0V0Z6Q0 A0A1W7R670 A0A2J7Q1Z2 A0A1W7R663 A0A2J7RKJ6 A0A2J7R668 A0A0A9Z683 A0A146LFV8 A0A2J7R8S8 A0A0V0RDY1 V5GAG2 A0A1B6E9D0 A0A0J7K0M9 A0A182GZK6 A0A1W7R6J6 A0A1W7R6K6 A0A0V0YAP7 A0A1Y1N3X2 A0A0V0YJB3 A0A0A9WQD4 A0A0V1HT08 A0A0V0YNF4 A0A0J7K5G8 K7J5Z3 A0A1W7R6D6 A0A0A9XVJ9 A0A0A9WPH5 A0A0S7KIW8 K7JBY9 A0A0K8U6S2 A0A0V1FXR9 A0A0A9WRE3 A0A0K8U956 A0A0A9XCL0 W8BYI7 A0A0A9WG89 A0A0A9WJ72 W8BFA2 A0A0J7K7B7 W8B528 W8AUM1 A0A0A1XSW5 A0A0A1WGJ2 W8ANX7 A0A0P6IXK4 K7JXM1 A0A034W7T5 A0A0A1X768 A0A2R6NZJ8 A0A0A1XLZ7 A0A182GNT8 A0A0J7K4F4 T1PKV2 W8BEX4 A0A2A4IIH8 A0A0V0V098 A0A251TN47 A0A0V1GS48 A0A131XNF9 B6V6Z8
Pubmed
EMBL
KQ460882
KPJ11482.1
GBHO01011746
GBHO01011744
GBHO01011742
JAG31858.1
+ More
JAG31860.1 JAG31862.1 LBMM01004924 KMQ91985.1 JYDJ01000877 KRX33198.1 NWSH01002292 PCG68657.1 PCG68658.1 GEZM01045162 JAV77889.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDH01000330 KRY26802.1 NWSH01000816 PCG74073.1 JYDT01000069 KRY86576.1 JYDJ01000808 KRX33413.1 GAKP01015352 JAC43600.1 JYDJ01000136 KRX42725.1 GEZM01011858 GEZM01011854 JAV93389.1 JYDK01000071 KRX78146.1 GBXI01011094 JAD03198.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 KQ460208 KPJ16606.1 GANO01000510 JAB59361.1 KQ460297 KPJ16236.1 KL367628 KFD61291.1 KRX78123.1 KL367553 KFD64618.1 KQ459875 KPJ19636.1 AGBW02009826 OWR49812.1 GBHO01026221 JAG17383.1 GEHC01000991 JAV46654.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 GEHC01000990 JAV46655.1 GFTR01008662 JAW07764.1 JYDQ01000379 KRY07987.1 GEHC01000989 JAV46656.1 NEVH01019375 PNF22597.1 GEHC01000988 JAV46657.1 NEVH01002725 PNF41351.1 NEVH01006980 PNF36330.1 GBHO01003630 JAG39974.1 GDHC01012180 JAQ06449.1 NEVH01006721 PNF37230.1 JYDL01000297 KRX12674.1 GALX01001351 JAB67115.1 GEDC01002834 JAS34464.1 LBMM01017716 KMQ83988.1 JXUM01099713 KQ564513 KXJ72145.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 JYDU01000039 KRX96809.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 JYDU01000010 KRY00136.1 GBHO01036544 JAG07060.1 JYDP01000032 KRZ13440.1 JYDU01000001 KRY01876.1 LBMM01013295 KMQ85718.1 GEHC01000965 JAV46680.1 GBHO01020706 JAG22898.1 GBHO01033237 JAG10367.1 GBYX01207955 JAO77249.1 AAZX01000798 GDHF01030254 JAI22060.1 JYDT01000024 KRY90099.1 GBHO01033235 JAG10369.1 GDHF01029191 JAI23123.1 GBHO01026223 JAG17381.1 GAMC01008189 JAB98366.1 GBHO01036147 JAG07457.1 GBHO01036148 JAG07456.1 GAMC01018156 JAB88399.1 LBMM01012669 KMQ86061.1 GAMC01018154 JAB88401.1 GAMC01018157 JAB88398.1 GBXI01000292 JAD14000.1 GBXI01016677 JAC97614.1 GAMC01020352 JAB86203.1 GDUN01000214 JAN95705.1 AAZX01023884 GAKP01008575 JAC50377.1 GBXI01007143 JAD07149.1 MLYV02000620 PSR81378.1 GBXI01002679 JAD11613.1 JXUM01077078 KQ562985 KXJ74708.1 LBMM01014442 KMQ85179.1 KA648745 AFP63374.1 GAMC01011082 JAB95473.1 NWVJ01000098 PCG17690.1 JYDN01000113 KRX56900.1 CM007899 OTG12557.1 JYDP01000350 KRZ01021.1 GEFH01000951 JAP67630.1 FJ238509 ACI62137.1
JAG31860.1 JAG31862.1 LBMM01004924 KMQ91985.1 JYDJ01000877 KRX33198.1 NWSH01002292 PCG68657.1 PCG68658.1 GEZM01045162 JAV77889.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDH01000330 KRY26802.1 NWSH01000816 PCG74073.1 JYDT01000069 KRY86576.1 JYDJ01000808 KRX33413.1 GAKP01015352 JAC43600.1 JYDJ01000136 KRX42725.1 GEZM01011858 GEZM01011854 JAV93389.1 JYDK01000071 KRX78146.1 GBXI01011094 JAD03198.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 KQ460208 KPJ16606.1 GANO01000510 JAB59361.1 KQ460297 KPJ16236.1 KL367628 KFD61291.1 KRX78123.1 KL367553 KFD64618.1 KQ459875 KPJ19636.1 AGBW02009826 OWR49812.1 GBHO01026221 JAG17383.1 GEHC01000991 JAV46654.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 GEHC01000990 JAV46655.1 GFTR01008662 JAW07764.1 JYDQ01000379 KRY07987.1 GEHC01000989 JAV46656.1 NEVH01019375 PNF22597.1 GEHC01000988 JAV46657.1 NEVH01002725 PNF41351.1 NEVH01006980 PNF36330.1 GBHO01003630 JAG39974.1 GDHC01012180 JAQ06449.1 NEVH01006721 PNF37230.1 JYDL01000297 KRX12674.1 GALX01001351 JAB67115.1 GEDC01002834 JAS34464.1 LBMM01017716 KMQ83988.1 JXUM01099713 KQ564513 KXJ72145.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 JYDU01000039 KRX96809.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 JYDU01000010 KRY00136.1 GBHO01036544 JAG07060.1 JYDP01000032 KRZ13440.1 JYDU01000001 KRY01876.1 LBMM01013295 KMQ85718.1 GEHC01000965 JAV46680.1 GBHO01020706 JAG22898.1 GBHO01033237 JAG10367.1 GBYX01207955 JAO77249.1 AAZX01000798 GDHF01030254 JAI22060.1 JYDT01000024 KRY90099.1 GBHO01033235 JAG10369.1 GDHF01029191 JAI23123.1 GBHO01026223 JAG17381.1 GAMC01008189 JAB98366.1 GBHO01036147 JAG07457.1 GBHO01036148 JAG07456.1 GAMC01018156 JAB88399.1 LBMM01012669 KMQ86061.1 GAMC01018154 JAB88401.1 GAMC01018157 JAB88398.1 GBXI01000292 JAD14000.1 GBXI01016677 JAC97614.1 GAMC01020352 JAB86203.1 GDUN01000214 JAN95705.1 AAZX01023884 GAKP01008575 JAC50377.1 GBXI01007143 JAD07149.1 MLYV02000620 PSR81378.1 GBXI01002679 JAD11613.1 JXUM01077078 KQ562985 KXJ74708.1 LBMM01014442 KMQ85179.1 KA648745 AFP63374.1 GAMC01011082 JAB95473.1 NWVJ01000098 PCG17690.1 JYDN01000113 KRX56900.1 CM007899 OTG12557.1 JYDP01000350 KRZ01021.1 GEFH01000951 JAP67630.1 FJ238509 ACI62137.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A0N1ICY1
A0A0A9YFM0
A0A0J7NHJ8
A0A0V0T2I4
A0A2A4J959
A0A1Y1LWI3
+ More
A0A0V1K1Y9 A0A0V1F2E6 E5SB91 A0A0V1APS7 A0A2A4JRI1 A0A0V1FKH2 A0A0V0T3I0 A0A034VP70 A0A0V0TUP0 A0A1Y1N6L8 A0A0V0WR33 A0A0A1WXQ3 A0A1Y1N668 A0A1Y1N932 A0A0N1IFR1 W4VRQ4 A0A194RJV9 A0A085MVP5 A0A0V0WQU6 A0A085N572 A0A194RU38 A0A212F7V4 A0A0A9XEY5 A0A1W7R657 W4VRP9 W4VRP2 A0A1W7R6H6 A0A224XA26 A0A0V0Z6Q0 A0A1W7R670 A0A2J7Q1Z2 A0A1W7R663 A0A2J7RKJ6 A0A2J7R668 A0A0A9Z683 A0A146LFV8 A0A2J7R8S8 A0A0V0RDY1 V5GAG2 A0A1B6E9D0 A0A0J7K0M9 A0A182GZK6 A0A1W7R6J6 A0A1W7R6K6 A0A0V0YAP7 A0A1Y1N3X2 A0A0V0YJB3 A0A0A9WQD4 A0A0V1HT08 A0A0V0YNF4 A0A0J7K5G8 K7J5Z3 A0A1W7R6D6 A0A0A9XVJ9 A0A0A9WPH5 A0A0S7KIW8 K7JBY9 A0A0K8U6S2 A0A0V1FXR9 A0A0A9WRE3 A0A0K8U956 A0A0A9XCL0 W8BYI7 A0A0A9WG89 A0A0A9WJ72 W8BFA2 A0A0J7K7B7 W8B528 W8AUM1 A0A0A1XSW5 A0A0A1WGJ2 W8ANX7 A0A0P6IXK4 K7JXM1 A0A034W7T5 A0A0A1X768 A0A2R6NZJ8 A0A0A1XLZ7 A0A182GNT8 A0A0J7K4F4 T1PKV2 W8BEX4 A0A2A4IIH8 A0A0V0V098 A0A251TN47 A0A0V1GS48 A0A131XNF9 B6V6Z8
A0A0V1K1Y9 A0A0V1F2E6 E5SB91 A0A0V1APS7 A0A2A4JRI1 A0A0V1FKH2 A0A0V0T3I0 A0A034VP70 A0A0V0TUP0 A0A1Y1N6L8 A0A0V0WR33 A0A0A1WXQ3 A0A1Y1N668 A0A1Y1N932 A0A0N1IFR1 W4VRQ4 A0A194RJV9 A0A085MVP5 A0A0V0WQU6 A0A085N572 A0A194RU38 A0A212F7V4 A0A0A9XEY5 A0A1W7R657 W4VRP9 W4VRP2 A0A1W7R6H6 A0A224XA26 A0A0V0Z6Q0 A0A1W7R670 A0A2J7Q1Z2 A0A1W7R663 A0A2J7RKJ6 A0A2J7R668 A0A0A9Z683 A0A146LFV8 A0A2J7R8S8 A0A0V0RDY1 V5GAG2 A0A1B6E9D0 A0A0J7K0M9 A0A182GZK6 A0A1W7R6J6 A0A1W7R6K6 A0A0V0YAP7 A0A1Y1N3X2 A0A0V0YJB3 A0A0A9WQD4 A0A0V1HT08 A0A0V0YNF4 A0A0J7K5G8 K7J5Z3 A0A1W7R6D6 A0A0A9XVJ9 A0A0A9WPH5 A0A0S7KIW8 K7JBY9 A0A0K8U6S2 A0A0V1FXR9 A0A0A9WRE3 A0A0K8U956 A0A0A9XCL0 W8BYI7 A0A0A9WG89 A0A0A9WJ72 W8BFA2 A0A0J7K7B7 W8B528 W8AUM1 A0A0A1XSW5 A0A0A1WGJ2 W8ANX7 A0A0P6IXK4 K7JXM1 A0A034W7T5 A0A0A1X768 A0A2R6NZJ8 A0A0A1XLZ7 A0A182GNT8 A0A0J7K4F4 T1PKV2 W8BEX4 A0A2A4IIH8 A0A0V0V098 A0A251TN47 A0A0V1GS48 A0A131XNF9 B6V6Z8
PDB
5T3A
E-value=0.0157647,
Score=91
Ontologies
KEGG
GO
PANTHER
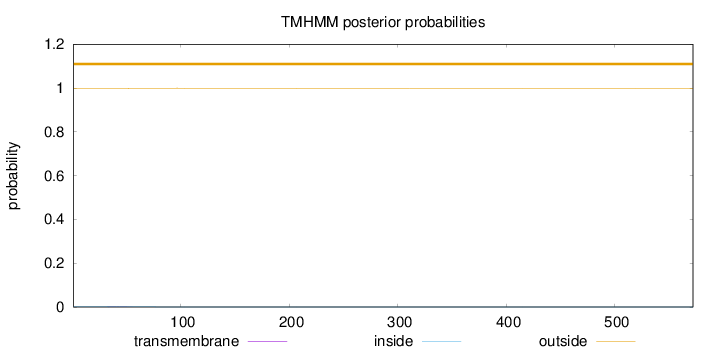
Topology
Length:
572
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0377400000000001
Exp number, first 60 AAs:
0.03228
Total prob of N-in:
0.00227
outside
1 - 572
Population Genetic Test Statistics
Pi
351.681561
Theta
156.592287
Tajima's D
3.94563
CLR
0.343657
CSRT
0.998800059997
Interpretation
Possibly Balancing Selection
