Gene
KWMTBOMO01288
Pre Gene Modal
BGIBMGA008872
Annotation
PREDICTED:_putative_inorganic_phosphate_cotransporter_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.974
Sequence
CDS
ATGAAATTCGAGAAAATACCATGTCGCTATATAATAGCATTGATGATATTCACCAGTTCATGCGCAAGTTTTATGATGCGCACGAATTTGAATGTGAATCTAGTCGCCATGGTTCCGGTTTCGAACAGTACTTATTCAGAAACCCAAACGACTTTAGATTGGACGCCATATCAGCGTGCAAATGTTCTATCAAGTCATTTATGGGGTGATATCTTGGCTTGTCTAATCGGTGGACCCATAGCGGAACGATGGGGTGCTCGCTATGTTGTTTTCTTTGGGATCCTGGTCAGCGCTGTATTGACAATGGCTACTCCTGCCGCTGCCAAGCACAGTTATATATCCATGTTGGTCGTTAGATTTATACTTGGATTTTTTACTGGATTTATTAATCCAGGGTGCCACGTTTTACTATCCAGATGGGCAGTAGCAACTGATAAAGGCAAATTCTCTGGAGCACTGATGGGTGGTAGCATAGGCATATTTATAGCTTGGAGTATATGCGGACAGCTCATCGAACCTTTTGGTTGGGAATGGGGATTCTATTTCTTTGGAATCGTCAGTTTTCCACTAATTGGTTTACTCTGGTTCTTGGTATACGATTCACCGGAAAAGCACCCTTACATTTCTGAAAAGGAGAAACAATACTTAACTGATAACATCCAAGTGCAACGTAAAAAAGTAAATGTTTCATAA
Protein
MKFEKIPCRYIIALMIFTSSCASFMMRTNLNVNLVAMVPVSNSTYSETQTTLDWTPYQRANVLSSHLWGDILACLIGGPIAERWGARYVVFFGILVSAVLTMATPAAAKHSYISMLVVRFILGFFTGFINPGCHVLLSRWAVATDKGKFSGALMGGSIGIFIAWSICGQLIEPFGWEWGFYFFGIVSFPLIGLLWFLVYDSPEKHPYISEKEKQYLTDNIQVQRKKVNVS
Summary
Uniprot
E3UVB6
H9JH75
A0A2H1WI55
A0A2W1BCG9
A0A212F8E0
A0A0L7KTA3
+ More
S4PDI4 A0A194R7P5 A0A194PUL2 A0A2H1WY07 A0A194PW78 A0A2W1BH44 H9JH72 A0A1J1I6K2 A0A0K8WJ76 A0A034VY00 A0A212ELN8 A0A0A1X1B5 A0A336LFL3 H9JH74 A0A336LRM6 A0A336MYY7 A0A0L0CL59 D6WVB6 A0A084WC96 A0A1I8Q857 A0A0M4EE35 Q28ZI2 B4GHQ6 B4IN92 B3MGS5 Q5R295 B4HQ44 Q5R274 A0A1Y1LHH1 Q494G1 A0A182PVF0 A0A1S4H4L7 A0A3B0JKY0 A0A1A9W0X1 B3NK44 A0A182HX26 B4PAN9 A0A2W1BAQ1 A0A182XA81 A0A1W4VI60 A0A182R8Z7 T1PC44 A0A182W889 A0A182MF47 A0A154P4C6 N6SZF2 U4U9S8 A0A182NQ00 A0A182K207 B4LPP1 B4MRX0 W8BYX5 Q7PRK0 A0A182YE46 E9IED7 A0A182QST5 A0A1L8DEC3 A0A1B6JRD3 B4KS25 A0A182F445 A0A026VZN4 A0A1A9XYA5 A0A195DXD1 A0A182S8R8 A0A1A9ZYI4 A0A151IB83 A0A1B0CFZ3 A0A1L8DER0 A0A1L8DEF1 A0A2A3EGE9 A0A1L8DEA4 B4JWG0 A0A1B0DPH9 A0A1B0DM98 Q16UY5 A0A1B6GXK0 B0XEM3 A0A1A9VB25 A0A0P4VXL1 T1I7Y0 R4G362 A0A2R7X080 A0A1L8DEH7 A0A1L8DEB6 A0A1B6CKE5 N6TMQ7 J3JTI0 J3JUH0 A0A146KJP9 A0A1B6KQM7 A0A1B0BUB9 A0A1L8DET0 A7RQH0 A0A1L8DEK7
S4PDI4 A0A194R7P5 A0A194PUL2 A0A2H1WY07 A0A194PW78 A0A2W1BH44 H9JH72 A0A1J1I6K2 A0A0K8WJ76 A0A034VY00 A0A212ELN8 A0A0A1X1B5 A0A336LFL3 H9JH74 A0A336LRM6 A0A336MYY7 A0A0L0CL59 D6WVB6 A0A084WC96 A0A1I8Q857 A0A0M4EE35 Q28ZI2 B4GHQ6 B4IN92 B3MGS5 Q5R295 B4HQ44 Q5R274 A0A1Y1LHH1 Q494G1 A0A182PVF0 A0A1S4H4L7 A0A3B0JKY0 A0A1A9W0X1 B3NK44 A0A182HX26 B4PAN9 A0A2W1BAQ1 A0A182XA81 A0A1W4VI60 A0A182R8Z7 T1PC44 A0A182W889 A0A182MF47 A0A154P4C6 N6SZF2 U4U9S8 A0A182NQ00 A0A182K207 B4LPP1 B4MRX0 W8BYX5 Q7PRK0 A0A182YE46 E9IED7 A0A182QST5 A0A1L8DEC3 A0A1B6JRD3 B4KS25 A0A182F445 A0A026VZN4 A0A1A9XYA5 A0A195DXD1 A0A182S8R8 A0A1A9ZYI4 A0A151IB83 A0A1B0CFZ3 A0A1L8DER0 A0A1L8DEF1 A0A2A3EGE9 A0A1L8DEA4 B4JWG0 A0A1B0DPH9 A0A1B0DM98 Q16UY5 A0A1B6GXK0 B0XEM3 A0A1A9VB25 A0A0P4VXL1 T1I7Y0 R4G362 A0A2R7X080 A0A1L8DEH7 A0A1L8DEB6 A0A1B6CKE5 N6TMQ7 J3JTI0 J3JUH0 A0A146KJP9 A0A1B6KQM7 A0A1B0BUB9 A0A1L8DET0 A7RQH0 A0A1L8DEK7
Pubmed
19121390
28756777
22118469
26227816
23622113
26354079
+ More
25348373 25830018 26108605 18362917 19820115 24438588 15632085 17994087 15729003 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 18057021 17550304 25315136 23537049 24495485 25244985 21282665 24508170 30249741 17510324 27129103 22516182 26823975 17615350
25348373 25830018 26108605 18362917 19820115 24438588 15632085 17994087 15729003 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 18057021 17550304 25315136 23537049 24495485 25244985 21282665 24508170 30249741 17510324 27129103 22516182 26823975 17615350
EMBL
HM593516
ADP37709.1
BABH01012813
BABH01012814
ODYU01008801
SOQ52717.1
+ More
KZ150406 PZC71007.1 AGBW02009746 OWR50000.1 JTDY01006140 KOB66249.1 GAIX01003661 JAA88899.1 KQ460597 KPJ13853.1 KQ459591 KPI97012.1 ODYU01011929 SOQ57955.1 KPI97014.1 PZC71003.1 BABH01012820 CVRI01000043 CRK95943.1 GDHF01001414 JAI50900.1 GAKP01011643 JAC47309.1 AGBW02014013 OWR42393.1 GBXI01009158 JAD05134.1 UFQS01004245 UFQT01004245 SSX16274.1 SSX35594.1 BABH01012815 SSX16272.1 SSX35592.1 UFQT01004233 SSX35584.1 JRES01000335 KNC32194.1 KQ971357 EFA08537.2 ATLV01022614 ATLV01022615 KE525334 KFB47840.1 CP012524 ALC41364.1 CM000071 EAL25631.1 CH479183 EDW36026.1 CH481762 EDW44601.1 CH902619 EDV36833.1 AB162349 CM000362 CM002911 BAD72899.1 EDX07882.1 KMY95223.1 CH480816 EDW48669.1 AB162351 BAD72917.1 GEZM01055166 GEZM01055165 GEZM01055164 GEZM01055163 JAV73102.1 AE013599 BT023815 AAF57499.1 AAZ66322.1 AAAB01008849 OUUW01000001 SPP73301.1 CH954179 EDV55279.1 KQS62211.1 APCN01005121 CM000158 EDW91430.1 KRJ99840.1 PZC71004.1 KA646259 AFP60888.1 AXCM01003116 KQ434813 KZC06795.1 APGK01050866 KB741178 ENN73149.1 KB632256 ERL90684.1 CH940648 EDW60279.1 CH963850 EDW74859.1 GAMC01008089 JAB98466.1 EAA07084.5 GL762576 EFZ21070.1 AXCN02002286 GFDF01009282 JAV04802.1 GECU01006054 JAT01653.1 CH933808 EDW10461.1 KK107648 QOIP01000008 EZA48344.1 RLU19777.1 KQ980167 KYN17372.1 KQ978112 KYM96920.1 AJWK01010573 GFDF01009270 JAV04814.1 GFDF01009319 JAV04765.1 KZ288262 PBC30372.1 GFDF01009302 JAV04782.1 CH916375 EDV98298.1 AJVK01018288 AJVK01006917 CH477609 EAT38350.1 GECZ01022282 GECZ01002621 JAS47487.1 JAS67148.1 DS232845 EDS26084.1 GDKW01002264 JAI54331.1 ACPB03012487 GAHY01001966 JAA75544.1 KK855891 PTY24275.1 GFDF01009307 JAV04777.1 GFDF01009298 JAV04786.1 GEDC01023334 JAS13964.1 APGK01058539 APGK01058540 KB741291 ENN70505.1 BT126537 AEE61501.1 BT126883 AEE61845.1 GDHC01021941 GDHC01010491 JAP96687.1 JAQ08138.1 GEBQ01026229 JAT13748.1 JXJN01020611 GFDF01009250 JAV04834.1 DS469528 EDO46366.1 GFDF01009269 JAV04815.1
KZ150406 PZC71007.1 AGBW02009746 OWR50000.1 JTDY01006140 KOB66249.1 GAIX01003661 JAA88899.1 KQ460597 KPJ13853.1 KQ459591 KPI97012.1 ODYU01011929 SOQ57955.1 KPI97014.1 PZC71003.1 BABH01012820 CVRI01000043 CRK95943.1 GDHF01001414 JAI50900.1 GAKP01011643 JAC47309.1 AGBW02014013 OWR42393.1 GBXI01009158 JAD05134.1 UFQS01004245 UFQT01004245 SSX16274.1 SSX35594.1 BABH01012815 SSX16272.1 SSX35592.1 UFQT01004233 SSX35584.1 JRES01000335 KNC32194.1 KQ971357 EFA08537.2 ATLV01022614 ATLV01022615 KE525334 KFB47840.1 CP012524 ALC41364.1 CM000071 EAL25631.1 CH479183 EDW36026.1 CH481762 EDW44601.1 CH902619 EDV36833.1 AB162349 CM000362 CM002911 BAD72899.1 EDX07882.1 KMY95223.1 CH480816 EDW48669.1 AB162351 BAD72917.1 GEZM01055166 GEZM01055165 GEZM01055164 GEZM01055163 JAV73102.1 AE013599 BT023815 AAF57499.1 AAZ66322.1 AAAB01008849 OUUW01000001 SPP73301.1 CH954179 EDV55279.1 KQS62211.1 APCN01005121 CM000158 EDW91430.1 KRJ99840.1 PZC71004.1 KA646259 AFP60888.1 AXCM01003116 KQ434813 KZC06795.1 APGK01050866 KB741178 ENN73149.1 KB632256 ERL90684.1 CH940648 EDW60279.1 CH963850 EDW74859.1 GAMC01008089 JAB98466.1 EAA07084.5 GL762576 EFZ21070.1 AXCN02002286 GFDF01009282 JAV04802.1 GECU01006054 JAT01653.1 CH933808 EDW10461.1 KK107648 QOIP01000008 EZA48344.1 RLU19777.1 KQ980167 KYN17372.1 KQ978112 KYM96920.1 AJWK01010573 GFDF01009270 JAV04814.1 GFDF01009319 JAV04765.1 KZ288262 PBC30372.1 GFDF01009302 JAV04782.1 CH916375 EDV98298.1 AJVK01018288 AJVK01006917 CH477609 EAT38350.1 GECZ01022282 GECZ01002621 JAS47487.1 JAS67148.1 DS232845 EDS26084.1 GDKW01002264 JAI54331.1 ACPB03012487 GAHY01001966 JAA75544.1 KK855891 PTY24275.1 GFDF01009307 JAV04777.1 GFDF01009298 JAV04786.1 GEDC01023334 JAS13964.1 APGK01058539 APGK01058540 KB741291 ENN70505.1 BT126537 AEE61501.1 BT126883 AEE61845.1 GDHC01021941 GDHC01010491 JAP96687.1 JAQ08138.1 GEBQ01026229 JAT13748.1 JXJN01020611 GFDF01009250 JAV04834.1 DS469528 EDO46366.1 GFDF01009269 JAV04815.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000053268
UP000183832
+ More
UP000037069 UP000007266 UP000030765 UP000095300 UP000092553 UP000001819 UP000008744 UP000001292 UP000007801 UP000000304 UP000000803 UP000075885 UP000268350 UP000091820 UP000008711 UP000075840 UP000002282 UP000076407 UP000192221 UP000075900 UP000095301 UP000075920 UP000075883 UP000076502 UP000019118 UP000030742 UP000075884 UP000075881 UP000008792 UP000007798 UP000007062 UP000076408 UP000075886 UP000009192 UP000069272 UP000053097 UP000279307 UP000092443 UP000078492 UP000075901 UP000092445 UP000078542 UP000092461 UP000242457 UP000001070 UP000092462 UP000008820 UP000002320 UP000078200 UP000015103 UP000092460 UP000001593
UP000037069 UP000007266 UP000030765 UP000095300 UP000092553 UP000001819 UP000008744 UP000001292 UP000007801 UP000000304 UP000000803 UP000075885 UP000268350 UP000091820 UP000008711 UP000075840 UP000002282 UP000076407 UP000192221 UP000075900 UP000095301 UP000075920 UP000075883 UP000076502 UP000019118 UP000030742 UP000075884 UP000075881 UP000008792 UP000007798 UP000007062 UP000076408 UP000075886 UP000009192 UP000069272 UP000053097 UP000279307 UP000092443 UP000078492 UP000075901 UP000092445 UP000078542 UP000092461 UP000242457 UP000001070 UP000092462 UP000008820 UP000002320 UP000078200 UP000015103 UP000092460 UP000001593
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
E3UVB6
H9JH75
A0A2H1WI55
A0A2W1BCG9
A0A212F8E0
A0A0L7KTA3
+ More
S4PDI4 A0A194R7P5 A0A194PUL2 A0A2H1WY07 A0A194PW78 A0A2W1BH44 H9JH72 A0A1J1I6K2 A0A0K8WJ76 A0A034VY00 A0A212ELN8 A0A0A1X1B5 A0A336LFL3 H9JH74 A0A336LRM6 A0A336MYY7 A0A0L0CL59 D6WVB6 A0A084WC96 A0A1I8Q857 A0A0M4EE35 Q28ZI2 B4GHQ6 B4IN92 B3MGS5 Q5R295 B4HQ44 Q5R274 A0A1Y1LHH1 Q494G1 A0A182PVF0 A0A1S4H4L7 A0A3B0JKY0 A0A1A9W0X1 B3NK44 A0A182HX26 B4PAN9 A0A2W1BAQ1 A0A182XA81 A0A1W4VI60 A0A182R8Z7 T1PC44 A0A182W889 A0A182MF47 A0A154P4C6 N6SZF2 U4U9S8 A0A182NQ00 A0A182K207 B4LPP1 B4MRX0 W8BYX5 Q7PRK0 A0A182YE46 E9IED7 A0A182QST5 A0A1L8DEC3 A0A1B6JRD3 B4KS25 A0A182F445 A0A026VZN4 A0A1A9XYA5 A0A195DXD1 A0A182S8R8 A0A1A9ZYI4 A0A151IB83 A0A1B0CFZ3 A0A1L8DER0 A0A1L8DEF1 A0A2A3EGE9 A0A1L8DEA4 B4JWG0 A0A1B0DPH9 A0A1B0DM98 Q16UY5 A0A1B6GXK0 B0XEM3 A0A1A9VB25 A0A0P4VXL1 T1I7Y0 R4G362 A0A2R7X080 A0A1L8DEH7 A0A1L8DEB6 A0A1B6CKE5 N6TMQ7 J3JTI0 J3JUH0 A0A146KJP9 A0A1B6KQM7 A0A1B0BUB9 A0A1L8DET0 A7RQH0 A0A1L8DEK7
S4PDI4 A0A194R7P5 A0A194PUL2 A0A2H1WY07 A0A194PW78 A0A2W1BH44 H9JH72 A0A1J1I6K2 A0A0K8WJ76 A0A034VY00 A0A212ELN8 A0A0A1X1B5 A0A336LFL3 H9JH74 A0A336LRM6 A0A336MYY7 A0A0L0CL59 D6WVB6 A0A084WC96 A0A1I8Q857 A0A0M4EE35 Q28ZI2 B4GHQ6 B4IN92 B3MGS5 Q5R295 B4HQ44 Q5R274 A0A1Y1LHH1 Q494G1 A0A182PVF0 A0A1S4H4L7 A0A3B0JKY0 A0A1A9W0X1 B3NK44 A0A182HX26 B4PAN9 A0A2W1BAQ1 A0A182XA81 A0A1W4VI60 A0A182R8Z7 T1PC44 A0A182W889 A0A182MF47 A0A154P4C6 N6SZF2 U4U9S8 A0A182NQ00 A0A182K207 B4LPP1 B4MRX0 W8BYX5 Q7PRK0 A0A182YE46 E9IED7 A0A182QST5 A0A1L8DEC3 A0A1B6JRD3 B4KS25 A0A182F445 A0A026VZN4 A0A1A9XYA5 A0A195DXD1 A0A182S8R8 A0A1A9ZYI4 A0A151IB83 A0A1B0CFZ3 A0A1L8DER0 A0A1L8DEF1 A0A2A3EGE9 A0A1L8DEA4 B4JWG0 A0A1B0DPH9 A0A1B0DM98 Q16UY5 A0A1B6GXK0 B0XEM3 A0A1A9VB25 A0A0P4VXL1 T1I7Y0 R4G362 A0A2R7X080 A0A1L8DEH7 A0A1L8DEB6 A0A1B6CKE5 N6TMQ7 J3JTI0 J3JUH0 A0A146KJP9 A0A1B6KQM7 A0A1B0BUB9 A0A1L8DET0 A7RQH0 A0A1L8DEK7
PDB
6E9O
E-value=3.30844e-06,
Score=118
Ontologies
GO
Topology
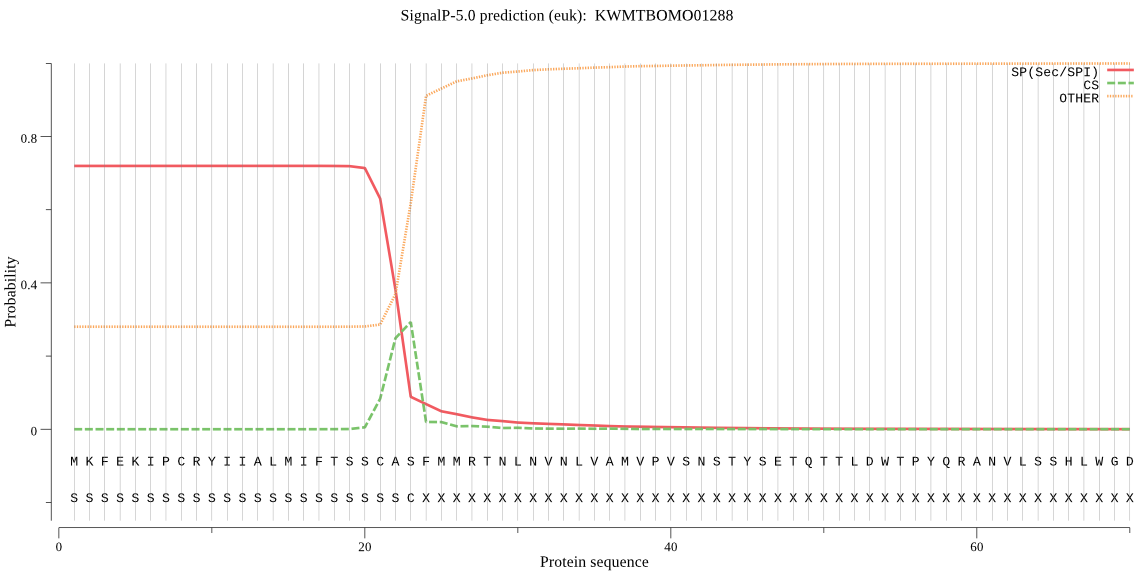
SignalP
Position: 1 - 23,
Likelihood: 0.719777
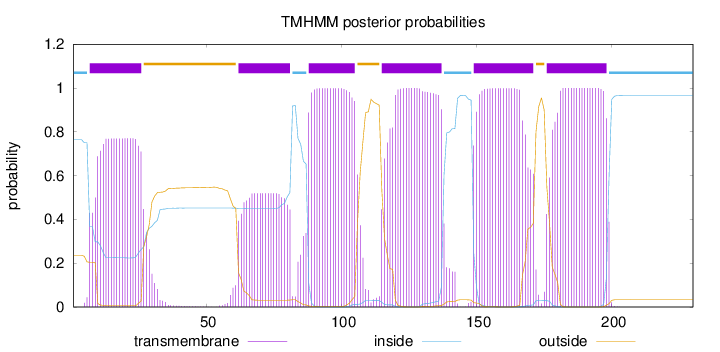
Length:
230
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
111.88078
Exp number, first 60 AAs:
15.97046
Total prob of N-in:
0.76382
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 61
TMhelix
62 - 81
inside
82 - 87
TMhelix
88 - 105
outside
106 - 114
TMhelix
115 - 137
inside
138 - 148
TMhelix
149 - 171
outside
172 - 175
TMhelix
176 - 198
inside
199 - 230
Population Genetic Test Statistics
Pi
220.943065
Theta
182.642073
Tajima's D
0.593552
CLR
0.354445
CSRT
0.547122643867807
Interpretation
Uncertain
