Gene
KWMTBOMO01286
Pre Gene Modal
BGIBMGA008870
Annotation
PREDICTED:_arginyl-tRNA--protein_transferase_1_[Papilio_polytes]
Full name
Arginyl-tRNA--protein transferase 1
Alternative Name
Arginine-tRNA--protein transferase 1
Location in the cell
Nuclear Reliability : 3.617
Sequence
CDS
ATGTTACGAAGTATTGTGGAATATTATGCAGAACACGAAAGATATAAATGTGGATACTGTAAAAACCTTGATACTAATTATAGTCATGGTCTGTGGACACATTCATTAACTGCACAGGATTATCAAGATCTCATTGACAGGGGTTGGAGAAGATCTGGAAAATATTGCTATAAACCCACATTGAATATTACTTGCTGCCCAATGTATACTATAAGATGCCAAGCACTAAAATTCAAAGCTACGAAATCCCAAAAGAAGATACTGAAGAAGTTCAACAAGCATCTCTTAGGACAAAACGAAAGTCACAGGAAGATGTCTACATGCAGTTCAAGTTCAGAAACATTGAATGAAACTTGTCAGAATGGAGCAGAACAGTTTATGCTGTGCACCAGAGATCATACTAAAATAAATGTAGATGATATTAAATTTATTGATGAGGATGATAGATTTCTTGGCGATAATGAGCAGCCAGTACAAAGAGGTGTACTAAAAAGTAATGAAAATACAAAAGAAACATGTAAAATTGATACAAATACAGGTAATTCTAGTCATGATCCCAACAGAGCCCCACTCAAGAAGGCAAAGTTATTAAGAAGAGAGAGAAAATTGGAAAAATTAAAACAGATGGGTATTGACACCACAACCATAAGTAATTCTAGTAAAACCAAGGAAAAACAATTAGAGGATTTCATAAATGAACTTCCAGATGATCCAAAACAAAAGTTGCAGATAAAATTAGTAAGAACAAACCCACCGAGCCCTGAATGGCAGGCGACCTCAAAGCAGACTCACGCTGTTTACTTAAAATATCAAACTACTGTACATGATGATGAACCTGACAAATGTACTGAAAGTAAATTTCATGACTTCTTACTCAACAGTCCTTTATTGGAAGAGTACAGTGCTAACGGGCCTCCCTGCGGCTACGGTTCTTTCCACCAACAGTATTGGCTTGACGGAAAGCTAATAGCCGTGGGGGTGATCGACATACTGCCGAAATGTGTTTCGTCTGTTTATTTCTTCTACGATCCAGATTATATGAATTTAACGCTCGGTACTTATGGAGCTCTGAGGGAAATCGAGTTCACAAGGTATCTGCACTCATTATGCCCGGATATTCAATATTACTATATGGGCTTTTACATACACTCTTGTCAAAAAATGAGGTACAAAGGGAACTTTCACCCCTCAGACTTGTTGTGCCCAGAGACTTACAAGTGGTTCCCGTTGAAGGATTGTATTCCTAAGTTGGAACTATCGCCGTATTCAAGATTGGACCCCGACATAGACAGTTTAGATACAAATCATCCTACTGAAGAGGATATTAATGATATACCGGTGTGGGTCAGTGGATCCGTGATGCTCTACAAAGTATTCAGGAGAAGCCTTATCTCGCGACGAGATGACAAAAGCGAAGTCCTCGAGTACGCGCGTTTTGTTGGAGCTAAAACAGCCAAAAGTCTGATACTAGTTAGATAA
Protein
MLRSIVEYYAEHERYKCGYCKNLDTNYSHGLWTHSLTAQDYQDLIDRGWRRSGKYCYKPTLNITCCPMYTIRCQALKFKATKSQKKILKKFNKHLLGQNESHRKMSTCSSSSETLNETCQNGAEQFMLCTRDHTKINVDDIKFIDEDDRFLGDNEQPVQRGVLKSNENTKETCKIDTNTGNSSHDPNRAPLKKAKLLRRERKLEKLKQMGIDTTTISNSSKTKEKQLEDFINELPDDPKQKLQIKLVRTNPPSPEWQATSKQTHAVYLKYQTTVHDDEPDKCTESKFHDFLLNSPLLEEYSANGPPCGYGSFHQQYWLDGKLIAVGVIDILPKCVSSVYFFYDPDYMNLTLGTYGALREIEFTRYLHSLCPDIQYYYMGFYIHSCQKMRYKGNFHPSDLLCPETYKWFPLKDCIPKLELSPYSRLDPDIDSLDTNHPTEEDINDIPVWVSGSVMLYKVFRRSLISRRDDKSEVLEYARFVGAKTAKSLILVR
Summary
Description
Involved in the post-translational conjugation of arginine to the N-terminal aspartate or glutamate of a protein. This arginylation is required for degradation of the protein via the ubiquitin pathway.
Involved in the post-translational conjugation of arginine to the N-terminal aspartate or glutamate of a protein. This arginylation is required for degradation of the protein via the ubiquitin pathway. Does not arginylate cysteine residues (By similarity).
Involved in the post-translational conjugation of arginine to the N-terminal aspartate or glutamate of a protein. This arginylation is required for degradation of the protein via the ubiquitin pathway. Does not arginylate cysteine residues (By similarity).
Catalytic Activity
a [protein] N-terminus + L-arginyl-tRNA(Arg) = H(+) + N-terminal L-arginyl-L-amino acid-[protein] + tRNA(Arg)
Subunit
Monomer. Interacts with LIAT1 (By similarity).
Similarity
Belongs to the R-transferase family.
Keywords
Acyltransferase
Alternative splicing
Complete proteome
Cytoplasm
Nucleus
Phosphoprotein
Reference proteome
Transferase
Ubl conjugation pathway
Feature
chain Arginyl-tRNA--protein transferase 1
splice variant In isoform ATE1-2.
splice variant In isoform ATE1-2.
Uniprot
H9JH73
A0A2W1BDJ6
A0A2H1WY74
A0A194R8M4
A0A212F6U4
T1HMM4
+ More
A0A3B4Z7E1 A0A3B4TIC5 A0A1B6IE36 A0A1W4YPX2 A0A1B6HRN9 A0A3P9IJP8 A0A1B6J6J3 A4IFW7 A0A2Y9DHP8 A0A3P9LVL7 H2MDC9 A0A3B5LGS1 A0A1S3N3J1 A0A3B4YMV4 A0A3Q0CTS5 A0A3P8UCD1 A0A3B4TIC8 A0A2D0QQ03 A0A218UQE9 A0A3Q0CU62 A0A3B4G4M3 A0A158P1B5 A0A3P8QSW9 A0A2Y9QUM4 A0A1W4YYW0 A0A2Y9DIC1 G8XPI0 A0A1W4XC30 G3UJP6 A0A2K5N398 A0A2R2ML81 A0A2I3MHV6 A0A2R9C5M4 A0A151NUW5 A0A2J8PBX4 K7D3K9 A0A2K6US96 A0A1B6EES1 A0A3B5L9B9 O95260 H0Y5C2 H9FP83 A0A2J8W6K1 A0A2J8W6K3 A0A3Q0CTX0 A0A2R2MKT5 G3IES1 A0A3B4BRJ7 A0A2Y9DIP7 A0A2K5EZF6 A0A286XY10 A0A151NU57 A0A287BLZ2 A0A3Q0CVD1 G3THT4 A0A1W4XM78 A0A2K5N3C8 A0A2I3LXR2 A0A3P8ZX36 A0A2I3T2K2 A0A286XSZ8 A0A2K6US92 O95260-2 A0A2J8W6L1 H9FP82 A0A3B3QF09 A0A2K6D6H1 U3JHH9 A0A2K5N3A8 A0A286ZNU5 A0A096NU10 A0A2R9BVX0 A0A2K6US81 A0A3B3YTM3 H0X1R8 A0A2I3SYG9 Q2PFX0 F7IL39 G1NWT5 A0A0S7I009 F5GXE4 H0WA95 A0A2K6NSP1 A0A2J8W6I3 Q5RB46 A0A2K6D6D1 A0A2K5EZH2 A0A3P8VZL5 R0JSL3 A0A2K5I2C7 U3IZM9 B4DR48 K9J5L5 A0A3B1KI34
A0A3B4Z7E1 A0A3B4TIC5 A0A1B6IE36 A0A1W4YPX2 A0A1B6HRN9 A0A3P9IJP8 A0A1B6J6J3 A4IFW7 A0A2Y9DHP8 A0A3P9LVL7 H2MDC9 A0A3B5LGS1 A0A1S3N3J1 A0A3B4YMV4 A0A3Q0CTS5 A0A3P8UCD1 A0A3B4TIC8 A0A2D0QQ03 A0A218UQE9 A0A3Q0CU62 A0A3B4G4M3 A0A158P1B5 A0A3P8QSW9 A0A2Y9QUM4 A0A1W4YYW0 A0A2Y9DIC1 G8XPI0 A0A1W4XC30 G3UJP6 A0A2K5N398 A0A2R2ML81 A0A2I3MHV6 A0A2R9C5M4 A0A151NUW5 A0A2J8PBX4 K7D3K9 A0A2K6US96 A0A1B6EES1 A0A3B5L9B9 O95260 H0Y5C2 H9FP83 A0A2J8W6K1 A0A2J8W6K3 A0A3Q0CTX0 A0A2R2MKT5 G3IES1 A0A3B4BRJ7 A0A2Y9DIP7 A0A2K5EZF6 A0A286XY10 A0A151NU57 A0A287BLZ2 A0A3Q0CVD1 G3THT4 A0A1W4XM78 A0A2K5N3C8 A0A2I3LXR2 A0A3P8ZX36 A0A2I3T2K2 A0A286XSZ8 A0A2K6US92 O95260-2 A0A2J8W6L1 H9FP82 A0A3B3QF09 A0A2K6D6H1 U3JHH9 A0A2K5N3A8 A0A286ZNU5 A0A096NU10 A0A2R9BVX0 A0A2K6US81 A0A3B3YTM3 H0X1R8 A0A2I3SYG9 Q2PFX0 F7IL39 G1NWT5 A0A0S7I009 F5GXE4 H0WA95 A0A2K6NSP1 A0A2J8W6I3 Q5RB46 A0A2K6D6D1 A0A2K5EZH2 A0A3P8VZL5 R0JSL3 A0A2K5I2C7 U3IZM9 B4DR48 K9J5L5 A0A3B1KI34
EC Number
2.3.2.8
Pubmed
EMBL
BABH01012816
BABH01012817
BABH01012818
KZ150406
PZC71006.1
ODYU01011929
+ More
SOQ57957.1 KQ460597 KPJ13854.1 AGBW02009964 OWR49466.1 ACPB03008511 GECU01022511 JAS85195.1 GECU01030363 JAS77343.1 GECU01012925 JAS94781.1 BC134811 AAI34812.1 MUZQ01000180 OWK55973.1 ADTU01006206 ADTU01006207 EU159112 ABX09996.1 AHZZ02036176 AJFE02090462 AJFE02090463 AJFE02090464 AJFE02090465 AJFE02090466 AJFE02090467 AJFE02090468 AJFE02090469 AJFE02090470 AJFE02090471 AKHW03001922 KYO40460.1 NBAG03000216 PNI81515.1 GABC01004145 GABD01006063 GABE01010460 JAA07193.1 JAA27037.1 JAA34279.1 PNI81511.1 GEDC01000944 JAS36354.1 AC025947 AL731542 AL731566 CH471066 BC022026 AF079098 AF079099 JU332687 JU473109 AFE76442.1 AFH29913.1 NDHI03003398 PNJ65393.1 PNJ65388.1 JH002256 EGW11696.1 AAKN02003000 AAKN02003001 AAKN02003002 AAKN02003003 AAKN02003004 KYO40461.1 AEMK02000096 DQIR01295596 DQIR01314590 HDC51074.1 AACZ04051230 GABC01004144 GABF01010145 GABD01006062 GABE01010459 JAA07194.1 JAA12000.1 JAA27038.1 JAA34280.1 PNI81514.1 PNJ65391.1 JU332686 JU473110 JV046051 AFE76441.1 AFH29914.1 AFI36122.1 AGTO01010916 AAQR03046576 AAQR03046577 AAQR03046578 AAQR03046579 AAQR03046580 PNI81509.1 AB220467 AAPE02056523 AAPE02056524 AAPE02056525 AAPE02056526 GBYX01434649 JAO46680.1 PNJ65386.1 CR858809 CAH91014.1 KB743226 EOB00182.1 ADON01083039 ADON01083040 ADON01083041 AK299101 BAG61160.1 GABZ01001516 JAA52009.1
SOQ57957.1 KQ460597 KPJ13854.1 AGBW02009964 OWR49466.1 ACPB03008511 GECU01022511 JAS85195.1 GECU01030363 JAS77343.1 GECU01012925 JAS94781.1 BC134811 AAI34812.1 MUZQ01000180 OWK55973.1 ADTU01006206 ADTU01006207 EU159112 ABX09996.1 AHZZ02036176 AJFE02090462 AJFE02090463 AJFE02090464 AJFE02090465 AJFE02090466 AJFE02090467 AJFE02090468 AJFE02090469 AJFE02090470 AJFE02090471 AKHW03001922 KYO40460.1 NBAG03000216 PNI81515.1 GABC01004145 GABD01006063 GABE01010460 JAA07193.1 JAA27037.1 JAA34279.1 PNI81511.1 GEDC01000944 JAS36354.1 AC025947 AL731542 AL731566 CH471066 BC022026 AF079098 AF079099 JU332687 JU473109 AFE76442.1 AFH29913.1 NDHI03003398 PNJ65393.1 PNJ65388.1 JH002256 EGW11696.1 AAKN02003000 AAKN02003001 AAKN02003002 AAKN02003003 AAKN02003004 KYO40461.1 AEMK02000096 DQIR01295596 DQIR01314590 HDC51074.1 AACZ04051230 GABC01004144 GABF01010145 GABD01006062 GABE01010459 JAA07194.1 JAA12000.1 JAA27038.1 JAA34280.1 PNI81514.1 PNJ65391.1 JU332686 JU473110 JV046051 AFE76441.1 AFH29914.1 AFI36122.1 AGTO01010916 AAQR03046576 AAQR03046577 AAQR03046578 AAQR03046579 AAQR03046580 PNI81509.1 AB220467 AAPE02056523 AAPE02056524 AAPE02056525 AAPE02056526 GBYX01434649 JAO46680.1 PNJ65386.1 CR858809 CAH91014.1 KB743226 EOB00182.1 ADON01083039 ADON01083040 ADON01083041 AK299101 BAG61160.1 GABZ01001516 JAA52009.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000015103
UP000261360
UP000261420
+ More
UP000192224 UP000265200 UP000248480 UP000265180 UP000001038 UP000261380 UP000087266 UP000189706 UP000265080 UP000221080 UP000197619 UP000261460 UP000005205 UP000265100 UP000192223 UP000007646 UP000233060 UP000085678 UP000028761 UP000240080 UP000050525 UP000233220 UP000005640 UP000001075 UP000261440 UP000233020 UP000005447 UP000008227 UP000265140 UP000002277 UP000261540 UP000233120 UP000016665 UP000261480 UP000005225 UP000233100 UP000008225 UP000001074 UP000233200 UP000265120 UP000233080 UP000016666 UP000018467
UP000192224 UP000265200 UP000248480 UP000265180 UP000001038 UP000261380 UP000087266 UP000189706 UP000265080 UP000221080 UP000197619 UP000261460 UP000005205 UP000265100 UP000192223 UP000007646 UP000233060 UP000085678 UP000028761 UP000240080 UP000050525 UP000233220 UP000005640 UP000001075 UP000261440 UP000233020 UP000005447 UP000008227 UP000265140 UP000002277 UP000261540 UP000233120 UP000016665 UP000261480 UP000005225 UP000233100 UP000008225 UP000001074 UP000233200 UP000265120 UP000233080 UP000016666 UP000018467
Interpro
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9JH73
A0A2W1BDJ6
A0A2H1WY74
A0A194R8M4
A0A212F6U4
T1HMM4
+ More
A0A3B4Z7E1 A0A3B4TIC5 A0A1B6IE36 A0A1W4YPX2 A0A1B6HRN9 A0A3P9IJP8 A0A1B6J6J3 A4IFW7 A0A2Y9DHP8 A0A3P9LVL7 H2MDC9 A0A3B5LGS1 A0A1S3N3J1 A0A3B4YMV4 A0A3Q0CTS5 A0A3P8UCD1 A0A3B4TIC8 A0A2D0QQ03 A0A218UQE9 A0A3Q0CU62 A0A3B4G4M3 A0A158P1B5 A0A3P8QSW9 A0A2Y9QUM4 A0A1W4YYW0 A0A2Y9DIC1 G8XPI0 A0A1W4XC30 G3UJP6 A0A2K5N398 A0A2R2ML81 A0A2I3MHV6 A0A2R9C5M4 A0A151NUW5 A0A2J8PBX4 K7D3K9 A0A2K6US96 A0A1B6EES1 A0A3B5L9B9 O95260 H0Y5C2 H9FP83 A0A2J8W6K1 A0A2J8W6K3 A0A3Q0CTX0 A0A2R2MKT5 G3IES1 A0A3B4BRJ7 A0A2Y9DIP7 A0A2K5EZF6 A0A286XY10 A0A151NU57 A0A287BLZ2 A0A3Q0CVD1 G3THT4 A0A1W4XM78 A0A2K5N3C8 A0A2I3LXR2 A0A3P8ZX36 A0A2I3T2K2 A0A286XSZ8 A0A2K6US92 O95260-2 A0A2J8W6L1 H9FP82 A0A3B3QF09 A0A2K6D6H1 U3JHH9 A0A2K5N3A8 A0A286ZNU5 A0A096NU10 A0A2R9BVX0 A0A2K6US81 A0A3B3YTM3 H0X1R8 A0A2I3SYG9 Q2PFX0 F7IL39 G1NWT5 A0A0S7I009 F5GXE4 H0WA95 A0A2K6NSP1 A0A2J8W6I3 Q5RB46 A0A2K6D6D1 A0A2K5EZH2 A0A3P8VZL5 R0JSL3 A0A2K5I2C7 U3IZM9 B4DR48 K9J5L5 A0A3B1KI34
A0A3B4Z7E1 A0A3B4TIC5 A0A1B6IE36 A0A1W4YPX2 A0A1B6HRN9 A0A3P9IJP8 A0A1B6J6J3 A4IFW7 A0A2Y9DHP8 A0A3P9LVL7 H2MDC9 A0A3B5LGS1 A0A1S3N3J1 A0A3B4YMV4 A0A3Q0CTS5 A0A3P8UCD1 A0A3B4TIC8 A0A2D0QQ03 A0A218UQE9 A0A3Q0CU62 A0A3B4G4M3 A0A158P1B5 A0A3P8QSW9 A0A2Y9QUM4 A0A1W4YYW0 A0A2Y9DIC1 G8XPI0 A0A1W4XC30 G3UJP6 A0A2K5N398 A0A2R2ML81 A0A2I3MHV6 A0A2R9C5M4 A0A151NUW5 A0A2J8PBX4 K7D3K9 A0A2K6US96 A0A1B6EES1 A0A3B5L9B9 O95260 H0Y5C2 H9FP83 A0A2J8W6K1 A0A2J8W6K3 A0A3Q0CTX0 A0A2R2MKT5 G3IES1 A0A3B4BRJ7 A0A2Y9DIP7 A0A2K5EZF6 A0A286XY10 A0A151NU57 A0A287BLZ2 A0A3Q0CVD1 G3THT4 A0A1W4XM78 A0A2K5N3C8 A0A2I3LXR2 A0A3P8ZX36 A0A2I3T2K2 A0A286XSZ8 A0A2K6US92 O95260-2 A0A2J8W6L1 H9FP82 A0A3B3QF09 A0A2K6D6H1 U3JHH9 A0A2K5N3A8 A0A286ZNU5 A0A096NU10 A0A2R9BVX0 A0A2K6US81 A0A3B3YTM3 H0X1R8 A0A2I3SYG9 Q2PFX0 F7IL39 G1NWT5 A0A0S7I009 F5GXE4 H0WA95 A0A2K6NSP1 A0A2J8W6I3 Q5RB46 A0A2K6D6D1 A0A2K5EZH2 A0A3P8VZL5 R0JSL3 A0A2K5I2C7 U3IZM9 B4DR48 K9J5L5 A0A3B1KI34
Ontologies
GO
PANTHER
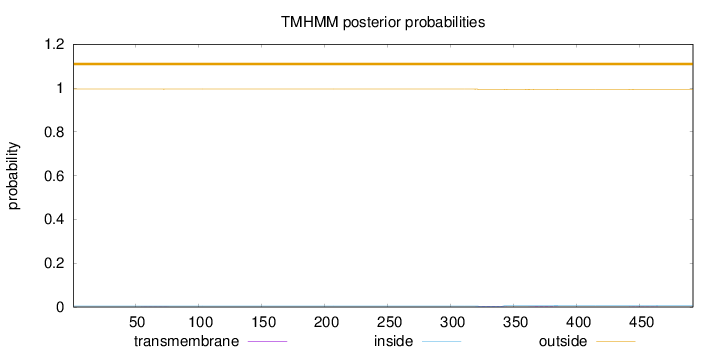
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11704
Exp number, first 60 AAs:
0.0018
Total prob of N-in:
0.00481
outside
1 - 492
Population Genetic Test Statistics
Pi
189.36687
Theta
164.071964
Tajima's D
0.84968
CLR
0.471102
CSRT
0.617469126543673
Interpretation
Uncertain
