Gene
KWMTBOMO01285
Pre Gene Modal
BGIBMGA008869
Annotation
PREDICTED:_uncharacterized_transporter_slc-17.2-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.906
Sequence
CDS
ATGGTGCCTCAAACGTCACACAATTCTTCTTCGACATCACAATGCGGAGCTAAAGACGACGTGGTCAGGAACACTAGTTTGCAGCACGTAGACATAGCTGATGTTAGAGAGGTGCCCAGACAGGTTCCAGGAGGTGCATCTTTTGATTGGACCGCTGATCAACAAGCACTTATCCTTGGCTCATACTTTTGGTGCTACCCGCTGACATCTTTGGCTGGAGGAATGGCTGCTGAGCGATGGGGTCCTCGATTAGTAGTCTTCGTGACTTCACTAGCAAGCGCAGTGCTAACTGCATTGAGTCCTGTGGCTTCATCCTGGAATTATCTGGCACTAGTTGTTATTAGGTTCTTCCTGGGATTTGCTTCAGGATTCATATATCCAGCCCTTCATGTGTTGGTAGCCCGGTGGGCTCCACCTGCGGAAAAAGGGAAGTTTGTTAGTGCCACGATGGGCGGTACATTAGGGACAGTTGTTACTTGGTCTTTGACTGGACCTCTCATAGAAAAGTTCGGCTGGGAGTCAGCGTTTTATGTGCCAGCCGCATTAACATTTGTTTGGTGCTTTTTCTGGTGGTACTTAGTTGCCGATACACCCTCAGAGCATCCAAGGATATCGGTAAAAGAGAAGAATTATATTTTAGATGCTCTAGGAGACAAGGTTAAAAAGTCAAAGGGCCTGCCGCCTTTTAAGAGCATTGTCAAGTCATTTCCCTTTCTGGCTATGATAGTTTTGCATTATGGAAACCTATGGGGATTGTACTTTATAATGACTGTTGGTCCAAAATTTGTGGCAAGCGTTTTAGGATTTGAATTATCAGCCGCGGGCGTAATATCCGCTTTACCATATCTCGCTAGGCTGGTGATGGCAACAATTTTTGGCTTTATTGGAGATTTTATATTAGCACGAAAATTAATGACAACGACGGTTATTCGAAAATTCTTCTGTCTTTTTTCACATATTTTGCCTGGATTATTGTTGCTCTGCTTGATTTATGCCGGATGTTCAACAACCCTATCAGTGGCTTTAATAACTATGTCAATGGGCTTCAACGGAGCTGCTACGTTAACAAATTTACAAAATCACCAAGACTTAGCTCCAAGTTATGCTGGAACTTTATATGGAATTGCTAATTGTATCGGTAGCACGGCTGGGTTTTTTACACCTATGATAACTGCATATTTCACAAGGAACGGAAACGGCTTCGAACAATGGAGACCAGTCTTTTATATAGGGGCGTCAGTATATGTTGTTTCTGCCGTATTTTTTATCTTCTTCGGCACCGGTAACATTCAAGATTGGAATTTTGCGGAAAATACAAAGGAAAATAAAGACGATAAAAACAACGACTTAAAAGAAATGAACGGGATAACGACACGAACCGGACTATTGGTGTCGTCATCTTGGCTATTTTCTATTTATCCAGGACCAATTCTATTGACTGTAGCTGTGGCTTTAACTGGAGCTGGAATGGATGTAACATTAGATTTATGCTATAATACTGCACTTCGGTGGAAAAATATTTGGTGTTCACATGCACTCGTATTATTGCTAAGTGGCATATTTTACTTAGTATGGGGCAAAACAGAAACCCAAGCATGGAATTTCAGCAGTCATACTCAGAGAAATCACCCAAAGAGTATTGTTAGAACGAATCCGTCGACTATGTCTAATATCACCGAAGTGGAGGAAGACGAGATAGCGAATACGTAA
Protein
MVPQTSHNSSSTSQCGAKDDVVRNTSLQHVDIADVREVPRQVPGGASFDWTADQQALILGSYFWCYPLTSLAGGMAAERWGPRLVVFVTSLASAVLTALSPVASSWNYLALVVIRFFLGFASGFIYPALHVLVARWAPPAEKGKFVSATMGGTLGTVVTWSLTGPLIEKFGWESAFYVPAALTFVWCFFWWYLVADTPSEHPRISVKEKNYILDALGDKVKKSKGLPPFKSIVKSFPFLAMIVLHYGNLWGLYFIMTVGPKFVASVLGFELSAAGVISALPYLARLVMATIFGFIGDFILARKLMTTTVIRKFFCLFSHILPGLLLLCLIYAGCSTTLSVALITMSMGFNGAATLTNLQNHQDLAPSYAGTLYGIANCIGSTAGFFTPMITAYFTRNGNGFEQWRPVFYIGASVYVVSAVFFIFFGTGNIQDWNFAENTKENKDDKNNDLKEMNGITTRTGLLVSSSWLFSIYPGPILLTVAVALTGAGMDVTLDLCYNTALRWKNIWCSHALVLLLSGIFYLVWGKTETQAWNFSSHTQRNHPKSIVRTNPSTMSNITEVEEDEIANT
Summary
Uniprot
H9JH72
A0A2W1BH44
A0A3S2LC91
A0A2H1WY07
S4PDI4
A0A194PW78
+ More
A0A212ELN8 A0A0L7KTA3 A0A194RD70 A0A2W1BAQ1 A0A2W1BCG9 A0A2H1WI55 A0A194R7P5 A0A194PUL2 A0A437AZH7 A0A212F8E0 H9JH74 D6WVB6 A0A182R8Z7 Q7PRK0 A0A1S4H4L7 A0A182W889 A0A182MF47 A0A1Y1LHH1 A0A182NQ00 A0A182S8R8 A0A182YE46 A0A1B0CFZ3 A0A1Q3FSI2 B0X3H7 B0XEM5 A0A182QST5 A0A1L8DE61 A0A1L8DEA7 A0A1L8DER0 A0A1L8DEH7 A0A1B0DPH9 A0A034VY00 A0A0K8WJ76 A0A182K207 U4U9S8 N6SZF2 Q494G1 B4PAN9 A0A182HX26 Q5R274 Q5R295 B3NK44 B4HQ44 A0A1W4VI60 W8BYX5 T1PC44 A0A0M4EE35 A0A182XA81 B4LPP1 B0X3H5 B4KS25 A0A182NPZ7 U5EJ09 A0A182PVE8 A0A182QSX6 A0A084WC98 W5JG06
A0A212ELN8 A0A0L7KTA3 A0A194RD70 A0A2W1BAQ1 A0A2W1BCG9 A0A2H1WI55 A0A194R7P5 A0A194PUL2 A0A437AZH7 A0A212F8E0 H9JH74 D6WVB6 A0A182R8Z7 Q7PRK0 A0A1S4H4L7 A0A182W889 A0A182MF47 A0A1Y1LHH1 A0A182NQ00 A0A182S8R8 A0A182YE46 A0A1B0CFZ3 A0A1Q3FSI2 B0X3H7 B0XEM5 A0A182QST5 A0A1L8DE61 A0A1L8DEA7 A0A1L8DER0 A0A1L8DEH7 A0A1B0DPH9 A0A034VY00 A0A0K8WJ76 A0A182K207 U4U9S8 N6SZF2 Q494G1 B4PAN9 A0A182HX26 Q5R274 Q5R295 B3NK44 B4HQ44 A0A1W4VI60 W8BYX5 T1PC44 A0A0M4EE35 A0A182XA81 B4LPP1 B0X3H5 B4KS25 A0A182NPZ7 U5EJ09 A0A182PVE8 A0A182QSX6 A0A084WC98 W5JG06
Pubmed
19121390
28756777
23622113
26354079
22118469
26227816
+ More
18362917 19820115 12364791 28004739 25244985 25348373 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 15729003 22936249 18057021 24495485 25315136 24438588 20920257 23761445
18362917 19820115 12364791 28004739 25244985 25348373 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 15729003 22936249 18057021 24495485 25315136 24438588 20920257 23761445
EMBL
BABH01012820
KZ150406
PZC71003.1
RSAL01000245
RVE43591.1
ODYU01011929
+ More
SOQ57955.1 GAIX01003661 JAA88899.1 KQ459591 KPI97014.1 AGBW02014013 OWR42393.1 JTDY01006140 KOB66249.1 KQ460597 KPJ13856.1 PZC71004.1 PZC71007.1 ODYU01008801 SOQ52717.1 KPJ13853.1 KPI97012.1 RVE43588.1 AGBW02009746 OWR50000.1 BABH01012815 KQ971357 EFA08537.2 AAAB01008849 EAA07084.5 AXCM01003116 GEZM01055166 GEZM01055165 GEZM01055164 GEZM01055163 JAV73102.1 AJWK01010573 GFDL01004609 JAV30436.1 DS232315 EDS39869.1 DS232845 EDS26086.1 AXCN02002286 GFDF01009331 JAV04753.1 GFDF01009308 JAV04776.1 GFDF01009270 JAV04814.1 GFDF01009307 JAV04777.1 AJVK01018288 GAKP01011643 JAC47309.1 GDHF01001414 JAI50900.1 KB632256 ERL90684.1 APGK01050866 KB741178 ENN73149.1 AE013599 BT023815 AAF57499.1 AAZ66322.1 CM000158 EDW91430.1 KRJ99840.1 APCN01005121 AB162351 BAD72917.1 AB162349 CM000362 CM002911 BAD72899.1 EDX07882.1 KMY95223.1 CH954179 EDV55279.1 KQS62211.1 CH480816 EDW48669.1 GAMC01008089 JAB98466.1 KA646259 AFP60888.1 CP012524 ALC41364.1 CH940648 EDW60279.1 EDS39867.1 CH933808 EDW10461.1 GANO01002416 JAB57455.1 ATLV01022615 KE525334 KFB47842.1 ADMH02001505 ETN62273.1
SOQ57955.1 GAIX01003661 JAA88899.1 KQ459591 KPI97014.1 AGBW02014013 OWR42393.1 JTDY01006140 KOB66249.1 KQ460597 KPJ13856.1 PZC71004.1 PZC71007.1 ODYU01008801 SOQ52717.1 KPJ13853.1 KPI97012.1 RVE43588.1 AGBW02009746 OWR50000.1 BABH01012815 KQ971357 EFA08537.2 AAAB01008849 EAA07084.5 AXCM01003116 GEZM01055166 GEZM01055165 GEZM01055164 GEZM01055163 JAV73102.1 AJWK01010573 GFDL01004609 JAV30436.1 DS232315 EDS39869.1 DS232845 EDS26086.1 AXCN02002286 GFDF01009331 JAV04753.1 GFDF01009308 JAV04776.1 GFDF01009270 JAV04814.1 GFDF01009307 JAV04777.1 AJVK01018288 GAKP01011643 JAC47309.1 GDHF01001414 JAI50900.1 KB632256 ERL90684.1 APGK01050866 KB741178 ENN73149.1 AE013599 BT023815 AAF57499.1 AAZ66322.1 CM000158 EDW91430.1 KRJ99840.1 APCN01005121 AB162351 BAD72917.1 AB162349 CM000362 CM002911 BAD72899.1 EDX07882.1 KMY95223.1 CH954179 EDV55279.1 KQS62211.1 CH480816 EDW48669.1 GAMC01008089 JAB98466.1 KA646259 AFP60888.1 CP012524 ALC41364.1 CH940648 EDW60279.1 EDS39867.1 CH933808 EDW10461.1 GANO01002416 JAB57455.1 ATLV01022615 KE525334 KFB47842.1 ADMH02001505 ETN62273.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000037510
UP000053240
+ More
UP000007266 UP000075900 UP000007062 UP000075920 UP000075883 UP000075884 UP000075901 UP000076408 UP000092461 UP000002320 UP000075886 UP000092462 UP000075881 UP000030742 UP000019118 UP000000803 UP000002282 UP000075840 UP000000304 UP000008711 UP000001292 UP000192221 UP000095301 UP000092553 UP000076407 UP000008792 UP000009192 UP000075885 UP000030765 UP000000673
UP000007266 UP000075900 UP000007062 UP000075920 UP000075883 UP000075884 UP000075901 UP000076408 UP000092461 UP000002320 UP000075886 UP000092462 UP000075881 UP000030742 UP000019118 UP000000803 UP000002282 UP000075840 UP000000304 UP000008711 UP000001292 UP000192221 UP000095301 UP000092553 UP000076407 UP000008792 UP000009192 UP000075885 UP000030765 UP000000673
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JH72
A0A2W1BH44
A0A3S2LC91
A0A2H1WY07
S4PDI4
A0A194PW78
+ More
A0A212ELN8 A0A0L7KTA3 A0A194RD70 A0A2W1BAQ1 A0A2W1BCG9 A0A2H1WI55 A0A194R7P5 A0A194PUL2 A0A437AZH7 A0A212F8E0 H9JH74 D6WVB6 A0A182R8Z7 Q7PRK0 A0A1S4H4L7 A0A182W889 A0A182MF47 A0A1Y1LHH1 A0A182NQ00 A0A182S8R8 A0A182YE46 A0A1B0CFZ3 A0A1Q3FSI2 B0X3H7 B0XEM5 A0A182QST5 A0A1L8DE61 A0A1L8DEA7 A0A1L8DER0 A0A1L8DEH7 A0A1B0DPH9 A0A034VY00 A0A0K8WJ76 A0A182K207 U4U9S8 N6SZF2 Q494G1 B4PAN9 A0A182HX26 Q5R274 Q5R295 B3NK44 B4HQ44 A0A1W4VI60 W8BYX5 T1PC44 A0A0M4EE35 A0A182XA81 B4LPP1 B0X3H5 B4KS25 A0A182NPZ7 U5EJ09 A0A182PVE8 A0A182QSX6 A0A084WC98 W5JG06
A0A212ELN8 A0A0L7KTA3 A0A194RD70 A0A2W1BAQ1 A0A2W1BCG9 A0A2H1WI55 A0A194R7P5 A0A194PUL2 A0A437AZH7 A0A212F8E0 H9JH74 D6WVB6 A0A182R8Z7 Q7PRK0 A0A1S4H4L7 A0A182W889 A0A182MF47 A0A1Y1LHH1 A0A182NQ00 A0A182S8R8 A0A182YE46 A0A1B0CFZ3 A0A1Q3FSI2 B0X3H7 B0XEM5 A0A182QST5 A0A1L8DE61 A0A1L8DEA7 A0A1L8DER0 A0A1L8DEH7 A0A1B0DPH9 A0A034VY00 A0A0K8WJ76 A0A182K207 U4U9S8 N6SZF2 Q494G1 B4PAN9 A0A182HX26 Q5R274 Q5R295 B3NK44 B4HQ44 A0A1W4VI60 W8BYX5 T1PC44 A0A0M4EE35 A0A182XA81 B4LPP1 B0X3H5 B4KS25 A0A182NPZ7 U5EJ09 A0A182PVE8 A0A182QSX6 A0A084WC98 W5JG06
PDB
6E9O
E-value=9.87185e-12,
Score=171
Ontologies
GO
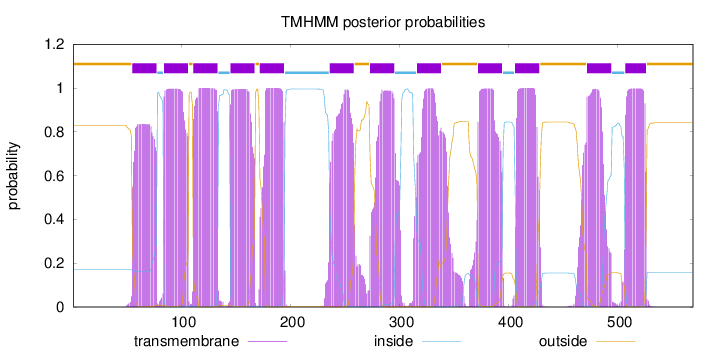
Topology
Length:
569
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
266.41811
Exp number, first 60 AAs:
4.50876
Total prob of N-in:
0.16961
outside
1 - 54
TMhelix
55 - 77
inside
78 - 83
TMhelix
84 - 106
outside
107 - 110
TMhelix
111 - 133
inside
134 - 144
TMhelix
145 - 167
outside
168 - 171
TMhelix
172 - 194
inside
195 - 235
TMhelix
236 - 258
outside
259 - 272
TMhelix
273 - 295
inside
296 - 315
TMhelix
316 - 338
outside
339 - 371
TMhelix
372 - 394
inside
395 - 405
TMhelix
406 - 428
outside
429 - 471
TMhelix
472 - 494
inside
495 - 506
TMhelix
507 - 526
outside
527 - 569
Population Genetic Test Statistics
Pi
258.624006
Theta
218.613487
Tajima's D
0.572537
CLR
0.335467
CSRT
0.533673316334183
Interpretation
Uncertain
