Gene
KWMTBOMO01281 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009018
Annotation
PREDICTED:_2-hydroxyphytanoyl-CoA_lyase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.476 Mitochondrial Reliability : 1.654
Sequence
CDS
ATGGGTATCGACGGAAATACAATCTTAGCTGAAAGTTTGAAGAAACAGGGTGTTGAATATGTATTTGGGATAGTTGGGATACCAGTCATTGAAACAGCAATGGCTTTCCAATCAGCTGGTCTAAAGTATATTGGTATGCGCAATGAACAAGCTGCCTGCTATGCTGCGCAAGCTACAGGCTATCTAACAGGTAAGCCAGGTGTTTGTTTAGTGGTATCAGGGCCGGGACTACTTCACTGCATCGGGGGAATGGCCAATGCTCAAGTAAACTGTTGGCCATTGGTTGTTATAGCTGGCTCGTGTCCTGAAGATCATGAAGCTATTGGTGGCTTCCAGGAATGGCCTCAGGTGGATTCTAGTCGCATGTACTGCAAGTATGCAGCCCGGCCGCCCTCCCCCCGTCTGATCCCCCAACACGTGGAGAAAGCTGTGCGACTTGCATTAGCCGGCCGTCCCGGAGTATCATATTTGGACATGCCCGGTACTCTGTTAATGGCAGAAGTGGACGAAGGCAAAGTACCACTAGACTACTACAGCGCAGACACGGTTAAACTGGCTCATCCCGATCCAGCGTTAGTAGATCAAGCGGCAGAATTACTTTCCAAAGCAGAAAGACCCCTGATCATAGTGGGCAAAGGCGCCGCATACGGCAAGGCCGAAGAGGCGATACGAAAACTAGTGGAAAACACAAAATTACCTTTCTTGCCCACACCAATGGGTAAAGGAGTGGTAGCTGACGAATCTGAGTACTGTGTATCAACTGCGCGTACACAAGCTCTACTGAAAGCTGATGTCATACTACTTCTTGGGGCTAGAATGAATTGGATGCTGCACTTTGGACAACCGCCGAGATATGCGCCCGACGTTAAAATTATTCAGGTCGAAATATCTCCTGAAGAATTTCACAACAGCAAGAAATCAGAATTAGCAGTACACTCAGACATTAGACCTTTTACAGAAGCTCTTGTAAAAAGGTTGTCCGAAAGGAAGTTCTCATTGCAACCTCAAAACAACAGTTGGTGGCAGGGACTGAAACAGAAACAAAAAGCAAACACCGAATTTGTAGAGGCTCAAGCAAGCAGCACTGCAGTACCACTGAACTACTACACTGTTTTCAAAACAGTTCAACAAGGCATTCCAAAAGATTCAATCATTGTCAGCGAAGGAGCGAATACCATGGATATTGGCCGAGGTTTATTGCTCAACAATCATCCGAGACACAGGCTAGATGCTGGTACATTTGGCACTATGGGGGTAGGACCTGGCTTCGCTATTGCAGCAGCAATGTGGTGCCGAGACTATGCACCTGGTAAACGTGTCATTTGTGTTGAAGGAGACTCTGCTTTTGGCTTCTCAGGCATGGAAATAGAGACAATGTTCCGGTACAAGTTACCTGTAATTATAATCATAGTAAACAATAATGGAATTTACAATGGGTTTGATAAAGAAGTAATGGCAGATATTCAATCTGGCGGCGATGTTACTCAGTGTACACCACCCACGGCTCTCAGTGGTGAAGTACGATACGAAAAGATGATGGAACTATTCGGCGAGACTGGTCATCTTTGTCGGACAGTCGAGGACATTAAAGAAGCAATTAAAACGGCCACAGCTGTTACTGATAAACCGAGTATCATCAACATCTTGATTGACCCACAGGCTAATAGGAAACCCCAAACGTTCAACTGGCTTACAGAATCCAAACTTTAA
Protein
MGIDGNTILAESLKKQGVEYVFGIVGIPVIETAMAFQSAGLKYIGMRNEQAACYAAQATGYLTGKPGVCLVVSGPGLLHCIGGMANAQVNCWPLVVIAGSCPEDHEAIGGFQEWPQVDSSRMYCKYAARPPSPRLIPQHVEKAVRLALAGRPGVSYLDMPGTLLMAEVDEGKVPLDYYSADTVKLAHPDPALVDQAAELLSKAERPLIIVGKGAAYGKAEEAIRKLVENTKLPFLPTPMGKGVVADESEYCVSTARTQALLKADVILLLGARMNWMLHFGQPPRYAPDVKIIQVEISPEEFHNSKKSELAVHSDIRPFTEALVKRLSERKFSLQPQNNSWWQGLKQKQKANTEFVEAQASSTAVPLNYYTVFKTVQQGIPKDSIIVSEGANTMDIGRGLLLNNHPRHRLDAGTFGTMGVGPGFAIAAAMWCRDYAPGKRVICVEGDSAFGFSGMEIETMFRYKLPVIIIIVNNNGIYNGFDKEVMADIQSGGDVTQCTPPTALSGEVRYEKMMELFGETGHLCRTVEDIKEAIKTATAVTDKPSIINILIDPQANRKPQTFNWLTESKL
Summary
Similarity
Belongs to the TPP enzyme family.
Uniprot
H9JHM0
Q2F5Z5
A0A2W1BJJ2
A0A2A4JNL1
A0A2A4JMR4
A0A212FNE1
+ More
A0A194Q0T3 A0A0N0PF82 A0A1B0DFY4 A0A2M4AKI8 W5JMJ4 A0A1Q3FEX5 A0A182YE44 A0A1L8DIW1 A0A182F447 B0X3H9 A0A034VXX4 A0A0K8VJQ2 B0XEM7 A0A182J1B2 A0A182HX25 A0A0Q9WEE0 A0A182H8A9 B4LPP2 W8BQ90 Q16FM3 A0A182NQ01 A0A1L8DIX1 A0A3B0JDI3 A0A1B0C8Q0 A0A182UM71 Q5R276 Q5R297 B4MRW7 B3NK68 Q16UY3 B4H4U9 A0A1I8PT35 B4HQ42 Q7K3B7 B4PAP1 A0A182U2B4 Q28X05 A0A1J1IP78 A0A182R8Z9 A0A0M4ECD2 B3MGS6 A0A1B0CFZ4 A0A1W4VGQ3 A0A182LN42 A0A182QZD9 A0A084WC95 B4JWF9 T1PDL1 A0A1I8N410 Q7QDR1 A0A182W888 A0A1L8EFW2 U5EKE4 A0A0L0CFN2 B4KS24 A0A1L8EHD5 A0A336LEU1 A0A023EUZ6 A0A182M381 A0A2J7Q906 A0A1A9W0X6 A0A1A9VB27 A0A026X0B9 A0A3L8DUG3 A0A158P1B6 A0A1A9ZYI5 A0A067R737 A0A1A9XYA6 A0A182XA82 E0VZ99 A0A195DL26 E2ATX6 A0A1B0G4P0 V5GVD7 E2C220 D6WWN7 A0A1B6DJS5 A0A1B6LZP6 A0A1B6F1Z5 A0A151K0C5 N6U7T1 A0A1B0BUC1 A0A151XGZ4 A0A1Y1KL51 A0A182K8W5 A0A2S2QZ66 A0A182PSC4 A0A0M9A9A6 A0A2S2P9X2 A0A2H8U0N6 A0A0K8TT28 A0A088A1S8 J9K0X7 A0A154P4W9
A0A194Q0T3 A0A0N0PF82 A0A1B0DFY4 A0A2M4AKI8 W5JMJ4 A0A1Q3FEX5 A0A182YE44 A0A1L8DIW1 A0A182F447 B0X3H9 A0A034VXX4 A0A0K8VJQ2 B0XEM7 A0A182J1B2 A0A182HX25 A0A0Q9WEE0 A0A182H8A9 B4LPP2 W8BQ90 Q16FM3 A0A182NQ01 A0A1L8DIX1 A0A3B0JDI3 A0A1B0C8Q0 A0A182UM71 Q5R276 Q5R297 B4MRW7 B3NK68 Q16UY3 B4H4U9 A0A1I8PT35 B4HQ42 Q7K3B7 B4PAP1 A0A182U2B4 Q28X05 A0A1J1IP78 A0A182R8Z9 A0A0M4ECD2 B3MGS6 A0A1B0CFZ4 A0A1W4VGQ3 A0A182LN42 A0A182QZD9 A0A084WC95 B4JWF9 T1PDL1 A0A1I8N410 Q7QDR1 A0A182W888 A0A1L8EFW2 U5EKE4 A0A0L0CFN2 B4KS24 A0A1L8EHD5 A0A336LEU1 A0A023EUZ6 A0A182M381 A0A2J7Q906 A0A1A9W0X6 A0A1A9VB27 A0A026X0B9 A0A3L8DUG3 A0A158P1B6 A0A1A9ZYI5 A0A067R737 A0A1A9XYA6 A0A182XA82 E0VZ99 A0A195DL26 E2ATX6 A0A1B0G4P0 V5GVD7 E2C220 D6WWN7 A0A1B6DJS5 A0A1B6LZP6 A0A1B6F1Z5 A0A151K0C5 N6U7T1 A0A1B0BUC1 A0A151XGZ4 A0A1Y1KL51 A0A182K8W5 A0A2S2QZ66 A0A182PSC4 A0A0M9A9A6 A0A2S2P9X2 A0A2H8U0N6 A0A0K8TT28 A0A088A1S8 J9K0X7 A0A154P4W9
Pubmed
19121390
28756777
22118469
26354079
20920257
23761445
+ More
25244985 25348373 17994087 26483478 24495485 17510324 15729003 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20966253 24438588 25315136 12364791 14747013 17210077 26108605 24945155 24508170 30249741 21347285 24845553 20566863 20798317 18362917 19820115 23537049 28004739 26369729
25244985 25348373 17994087 26483478 24495485 17510324 15729003 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20966253 24438588 25315136 12364791 14747013 17210077 26108605 24945155 24508170 30249741 21347285 24845553 20566863 20798317 18362917 19820115 23537049 28004739 26369729
EMBL
BABH01012825
BABH01012826
BABH01012827
DQ311277
ABD36222.1
KZ150082
+ More
PZC73834.1 NWSH01000967 PCG73366.1 PCG73365.1 AGBW02005912 OWR55255.1 KQ459591 KPI97015.1 LADJ01009455 KPJ20931.1 AJVK01059120 GGFK01007970 MBW41291.1 ADMH02000978 ETN64528.1 GFDL01008947 JAV26098.1 GFDF01007691 JAV06393.1 DS232315 EDS39871.1 GAKP01010791 JAC48161.1 GDHF01013212 JAI39102.1 DS232845 EDS26088.1 APCN01005120 CH940648 KRF79342.1 JXUM01030206 KQ560877 KXJ80509.1 EDW60280.1 GAMC01014856 JAB91699.1 CH478411 EAT33038.1 GFDF01007692 JAV06392.1 OUUW01000001 SPP73300.1 AJWK01001234 AB162351 BAD72915.1 AB162349 CM002911 BAD72897.1 KMY95221.1 CH963850 EDW74856.2 CH954179 EDV55277.1 CH477609 EAT38352.1 CH479210 EDW32785.1 CH480816 EDW48667.1 AE013599 AY058683 AAF57501.1 AAL13912.1 CM000158 EDW91432.1 CM000071 EAL26511.1 CVRI01000057 CRL01968.1 CP012524 ALC41365.1 CH902619 EDV36834.1 AJWK01010573 AXCN02002286 ATLV01022612 KE525334 KFB47839.1 CH916375 EDV98297.1 KA646846 AFP61475.1 AAAB01008849 EAA07115.3 GFDG01001310 JAV17489.1 GANO01001843 JAB58028.1 JRES01000462 KNC31012.1 CH933808 EDW10460.1 GFDG01000657 JAV18142.1 UFQS01003096 UFQT01003096 SSX15181.1 SSX34557.1 GAPW01000781 JAC12817.1 AXCM01003116 AXCM01003117 NEVH01016943 PNF25061.1 KK107046 EZA61740.1 QOIP01000004 RLU23932.1 ADTU01006210 ADTU01006211 ADTU01006212 KK852699 KDR18238.1 DS235851 EEB18705.1 KQ980762 KYN13585.1 GL442747 EFN63107.1 CCAG010021607 GALX01004268 JAB64198.1 GL452067 EFN77971.1 KQ971361 EFA08110.1 GEDC01011393 JAS25905.1 GEBQ01010867 JAT29110.1 GECZ01025463 JAS44306.1 KQ981301 KYN43039.1 APGK01036344 KB740939 ENN77730.1 JXJN01020611 KQ982138 KYQ59674.1 GEZM01083860 JAV60970.1 GGMS01013855 MBY83058.1 KQ435710 KOX79840.1 GGMR01013634 MBY26253.1 GFXV01007934 MBW19739.1 GDAI01000084 JAI17519.1 ABLF02013065 KQ434819 KZC06969.1
PZC73834.1 NWSH01000967 PCG73366.1 PCG73365.1 AGBW02005912 OWR55255.1 KQ459591 KPI97015.1 LADJ01009455 KPJ20931.1 AJVK01059120 GGFK01007970 MBW41291.1 ADMH02000978 ETN64528.1 GFDL01008947 JAV26098.1 GFDF01007691 JAV06393.1 DS232315 EDS39871.1 GAKP01010791 JAC48161.1 GDHF01013212 JAI39102.1 DS232845 EDS26088.1 APCN01005120 CH940648 KRF79342.1 JXUM01030206 KQ560877 KXJ80509.1 EDW60280.1 GAMC01014856 JAB91699.1 CH478411 EAT33038.1 GFDF01007692 JAV06392.1 OUUW01000001 SPP73300.1 AJWK01001234 AB162351 BAD72915.1 AB162349 CM002911 BAD72897.1 KMY95221.1 CH963850 EDW74856.2 CH954179 EDV55277.1 CH477609 EAT38352.1 CH479210 EDW32785.1 CH480816 EDW48667.1 AE013599 AY058683 AAF57501.1 AAL13912.1 CM000158 EDW91432.1 CM000071 EAL26511.1 CVRI01000057 CRL01968.1 CP012524 ALC41365.1 CH902619 EDV36834.1 AJWK01010573 AXCN02002286 ATLV01022612 KE525334 KFB47839.1 CH916375 EDV98297.1 KA646846 AFP61475.1 AAAB01008849 EAA07115.3 GFDG01001310 JAV17489.1 GANO01001843 JAB58028.1 JRES01000462 KNC31012.1 CH933808 EDW10460.1 GFDG01000657 JAV18142.1 UFQS01003096 UFQT01003096 SSX15181.1 SSX34557.1 GAPW01000781 JAC12817.1 AXCM01003116 AXCM01003117 NEVH01016943 PNF25061.1 KK107046 EZA61740.1 QOIP01000004 RLU23932.1 ADTU01006210 ADTU01006211 ADTU01006212 KK852699 KDR18238.1 DS235851 EEB18705.1 KQ980762 KYN13585.1 GL442747 EFN63107.1 CCAG010021607 GALX01004268 JAB64198.1 GL452067 EFN77971.1 KQ971361 EFA08110.1 GEDC01011393 JAS25905.1 GEBQ01010867 JAT29110.1 GECZ01025463 JAS44306.1 KQ981301 KYN43039.1 APGK01036344 KB740939 ENN77730.1 JXJN01020611 KQ982138 KYQ59674.1 GEZM01083860 JAV60970.1 GGMS01013855 MBY83058.1 KQ435710 KOX79840.1 GGMR01013634 MBY26253.1 GFXV01007934 MBW19739.1 GDAI01000084 JAI17519.1 ABLF02013065 KQ434819 KZC06969.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000092462
+ More
UP000000673 UP000076408 UP000069272 UP000002320 UP000075880 UP000075840 UP000008792 UP000069940 UP000249989 UP000008820 UP000075884 UP000268350 UP000092461 UP000075903 UP000007798 UP000008711 UP000008744 UP000095300 UP000001292 UP000000803 UP000002282 UP000075902 UP000001819 UP000183832 UP000075900 UP000092553 UP000007801 UP000192221 UP000075882 UP000075886 UP000030765 UP000001070 UP000095301 UP000007062 UP000075920 UP000037069 UP000009192 UP000075883 UP000235965 UP000091820 UP000078200 UP000053097 UP000279307 UP000005205 UP000092445 UP000027135 UP000092443 UP000076407 UP000009046 UP000078492 UP000000311 UP000092444 UP000008237 UP000007266 UP000078541 UP000019118 UP000092460 UP000075809 UP000075881 UP000075885 UP000053105 UP000005203 UP000007819 UP000076502
UP000000673 UP000076408 UP000069272 UP000002320 UP000075880 UP000075840 UP000008792 UP000069940 UP000249989 UP000008820 UP000075884 UP000268350 UP000092461 UP000075903 UP000007798 UP000008711 UP000008744 UP000095300 UP000001292 UP000000803 UP000002282 UP000075902 UP000001819 UP000183832 UP000075900 UP000092553 UP000007801 UP000192221 UP000075882 UP000075886 UP000030765 UP000001070 UP000095301 UP000007062 UP000075920 UP000037069 UP000009192 UP000075883 UP000235965 UP000091820 UP000078200 UP000053097 UP000279307 UP000005205 UP000092445 UP000027135 UP000092443 UP000076407 UP000009046 UP000078492 UP000000311 UP000092444 UP000008237 UP000007266 UP000078541 UP000019118 UP000092460 UP000075809 UP000075881 UP000075885 UP000053105 UP000005203 UP000007819 UP000076502
Interpro
ProteinModelPortal
H9JHM0
Q2F5Z5
A0A2W1BJJ2
A0A2A4JNL1
A0A2A4JMR4
A0A212FNE1
+ More
A0A194Q0T3 A0A0N0PF82 A0A1B0DFY4 A0A2M4AKI8 W5JMJ4 A0A1Q3FEX5 A0A182YE44 A0A1L8DIW1 A0A182F447 B0X3H9 A0A034VXX4 A0A0K8VJQ2 B0XEM7 A0A182J1B2 A0A182HX25 A0A0Q9WEE0 A0A182H8A9 B4LPP2 W8BQ90 Q16FM3 A0A182NQ01 A0A1L8DIX1 A0A3B0JDI3 A0A1B0C8Q0 A0A182UM71 Q5R276 Q5R297 B4MRW7 B3NK68 Q16UY3 B4H4U9 A0A1I8PT35 B4HQ42 Q7K3B7 B4PAP1 A0A182U2B4 Q28X05 A0A1J1IP78 A0A182R8Z9 A0A0M4ECD2 B3MGS6 A0A1B0CFZ4 A0A1W4VGQ3 A0A182LN42 A0A182QZD9 A0A084WC95 B4JWF9 T1PDL1 A0A1I8N410 Q7QDR1 A0A182W888 A0A1L8EFW2 U5EKE4 A0A0L0CFN2 B4KS24 A0A1L8EHD5 A0A336LEU1 A0A023EUZ6 A0A182M381 A0A2J7Q906 A0A1A9W0X6 A0A1A9VB27 A0A026X0B9 A0A3L8DUG3 A0A158P1B6 A0A1A9ZYI5 A0A067R737 A0A1A9XYA6 A0A182XA82 E0VZ99 A0A195DL26 E2ATX6 A0A1B0G4P0 V5GVD7 E2C220 D6WWN7 A0A1B6DJS5 A0A1B6LZP6 A0A1B6F1Z5 A0A151K0C5 N6U7T1 A0A1B0BUC1 A0A151XGZ4 A0A1Y1KL51 A0A182K8W5 A0A2S2QZ66 A0A182PSC4 A0A0M9A9A6 A0A2S2P9X2 A0A2H8U0N6 A0A0K8TT28 A0A088A1S8 J9K0X7 A0A154P4W9
A0A194Q0T3 A0A0N0PF82 A0A1B0DFY4 A0A2M4AKI8 W5JMJ4 A0A1Q3FEX5 A0A182YE44 A0A1L8DIW1 A0A182F447 B0X3H9 A0A034VXX4 A0A0K8VJQ2 B0XEM7 A0A182J1B2 A0A182HX25 A0A0Q9WEE0 A0A182H8A9 B4LPP2 W8BQ90 Q16FM3 A0A182NQ01 A0A1L8DIX1 A0A3B0JDI3 A0A1B0C8Q0 A0A182UM71 Q5R276 Q5R297 B4MRW7 B3NK68 Q16UY3 B4H4U9 A0A1I8PT35 B4HQ42 Q7K3B7 B4PAP1 A0A182U2B4 Q28X05 A0A1J1IP78 A0A182R8Z9 A0A0M4ECD2 B3MGS6 A0A1B0CFZ4 A0A1W4VGQ3 A0A182LN42 A0A182QZD9 A0A084WC95 B4JWF9 T1PDL1 A0A1I8N410 Q7QDR1 A0A182W888 A0A1L8EFW2 U5EKE4 A0A0L0CFN2 B4KS24 A0A1L8EHD5 A0A336LEU1 A0A023EUZ6 A0A182M381 A0A2J7Q906 A0A1A9W0X6 A0A1A9VB27 A0A026X0B9 A0A3L8DUG3 A0A158P1B6 A0A1A9ZYI5 A0A067R737 A0A1A9XYA6 A0A182XA82 E0VZ99 A0A195DL26 E2ATX6 A0A1B0G4P0 V5GVD7 E2C220 D6WWN7 A0A1B6DJS5 A0A1B6LZP6 A0A1B6F1Z5 A0A151K0C5 N6U7T1 A0A1B0BUC1 A0A151XGZ4 A0A1Y1KL51 A0A182K8W5 A0A2S2QZ66 A0A182PSC4 A0A0M9A9A6 A0A2S2P9X2 A0A2H8U0N6 A0A0K8TT28 A0A088A1S8 J9K0X7 A0A154P4W9
PDB
2Q29
E-value=7.39175e-89,
Score=836
Ontologies
GO
Topology
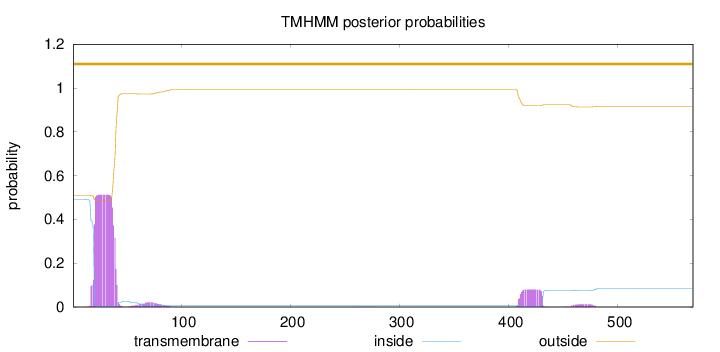
Length:
569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.71742
Exp number, first 60 AAs:
10.38022
Total prob of N-in:
0.49077
POSSIBLE N-term signal
sequence
outside
1 - 569
Population Genetic Test Statistics
Pi
200.260952
Theta
186.969762
Tajima's D
0.184825
CLR
1.605919
CSRT
0.420628968551572
Interpretation
Uncertain
