Gene
KWMTBOMO01274 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009021
Annotation
PREDICTED:_calcyclin-binding_protein_[Amyelois_transitella]
Full name
Calcyclin-binding protein
Location in the cell
Nuclear Reliability : 3.504
Sequence
CDS
ATGTCAGAAGCAAAAATACAAGAGATAAGAAGCGATATCGAAGAGATCAATGATCTGTTAAAGCAAGCTAAACGTAAGAAAGTACAGGATTTACTCTCCTTGGAGATCAGAAAACTTGAGACAGAACTAATAACATTAAAAGAAAAGGAAACTTTGACGCCCATGGAAGTATCACCAATACCCACAACAAGTACATCAGCTCCAGTGCAAAAGAAGTATCAAGTCAAATTGAATGTATATGGTTGGGACCAGTCGGATAAATTTGTTAAGGTATTTGTTGAACTAAAGAATGTTCATACACTACCAAAGGAACAGGTATATTGTAAATTGACAGATAAATCAATGGAGCTGCATGTGGACAATCTGGAAAATAAAGATTACTTGCTTGTTATTAACAAGTTATTGGAACCTATAAATGTGGCAGACAGTCATTGGAAACAAAAAACAGATAAGGTAGTCATATTCCTTGCAAAATCTAATCCAAACACAACATGGTCTCATATGACTGAAATTGAAAAAAAATTTGAAGACCAACGTAATAACCGCTTGAAACCTGCCGAAACTGACAAGAAAGATCCACAAGATTCCATAATGAGTCTCATGAAGAATATGTATGAAACTGGTGATGATGAAATGAAGAGAATGATCTCCAAGGCTTGGTATGAAGGACAGCAGCGTAAGAAAACTGATATAATGGATTTATAA
Protein
MSEAKIQEIRSDIEEINDLLKQAKRKKVQDLLSLEIRKLETELITLKEKETLTPMEVSPIPTTSTSAPVQKKYQVKLNVYGWDQSDKFVKVFVELKNVHTLPKEQVYCKLTDKSMELHVDNLENKDYLLVINKLLEPINVADSHWKQKTDKVVIFLAKSNPNTTWSHMTEIEKKFEDQRNNRLKPAETDKKDPQDSIMSLMKNMYETGDDEMKRMISKAWYEGQQRKKTDIMDL
Summary
Description
May be involved in calcium-dependent ubiquitination and subsequent proteasomal degradation of target proteins. Probably serves as a molecular bridge in ubiquitin E3 complexes. Participates in the ubiquitin-mediated degradation of beta-catenin (CTNNB1) (By similarity).
Subunit
Monomer or homodimer. Component of some large E3 complex at least composed of UBE2D1, SIAH1, CACYBP/SIP, SKP1, APC and TBL1X. Interacts directly with SIAH1, SIAH2 and SKP1. Interacts with proteins of the S100 family S100A1, S100A6, S100B, S100P and S100A12 in a calcium-dependent manner (By similarity).
Keywords
Acetylation
Complete proteome
Cytoplasm
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation pathway
Feature
chain Calcyclin-binding protein
Uniprot
H9JHM3
A0A194PUM2
A0A194R8N4
A0A2H1X061
A0A2W1BMA2
A0A2A4IV95
+ More
A0A212EHF9 S4PE51 A0A1B6MMV3 A0A1B6H7D3 A0A1W4WIL7 A0A2J7QUK2 A0A067R418 A0A2P8YCA2 D7GXT5 A0A0J7MRE2 A0A1B6CCB6 E2BSE3 K7IRE0 V5H2G8 A0A232F303 A0A158NH28 E2AA03 A0A195D1F5 A0A088AK08 I6LNT1 A0A154PPW2 A0A069DR20 A0A0V0G4V8 A0A151XDB4 A0A0P4VL48 R4G3G8 A0A0L7R511 F4WSV6 A0A023F876 A0A0P5A3G8 A0A2R7W062 A0A0P5XMP6 A0A0P4ZI35 A0A1Y1M5A7 A0A182F7Z1 A0A3L8DN99 A0A1L8DLP9 A0A0P5R797 A0A023EKU1 A0A1L8DLZ4 W5JAT2 U5ENM9 A0A310SQ90 Q16JJ2 N6TWA1 A0A0P5BSU5 E9IMS7 A0A182G1Q6 T1E7K4 A0A084W9U3 A0A182XW24 B0XGQ8 A0A0P5XQB8 A0A1A9Z3X0 A0A0L0C1U4 A0A1Q3F5T1 K9IZ15 A0A0M9AA74 A0A1B0D1S2 A0A1B0FNW9 A0A182XGG9 A0A182Q7R6 A0A182N6A4 A0A250YCT2 L5KYI4 A0A182LYA8 A0A2M4AR34 A0A182PNB7 E9GDE7 A0A2C9GRK4 Q7PYU2 A0A1A9VWX7 H2ZZ02 A0A182UY46 A0A1A9XYJ1 A0A3B3QP14 C1BVY9 A0A2K6MEQ0 A0A182LQY2 S7MVZ0 A0A182UAE3 A0A1B0ASR9 A0A1B0CX15 G3TH46 Q7ZWU4 A0A1U7USE1 M3W9V3 A0A2Y9GLN5 A0A2U3XP45 A0A2U3X389 Q3T168 A0A182IN24 A0A182K5T1 A0A2U3V766
A0A212EHF9 S4PE51 A0A1B6MMV3 A0A1B6H7D3 A0A1W4WIL7 A0A2J7QUK2 A0A067R418 A0A2P8YCA2 D7GXT5 A0A0J7MRE2 A0A1B6CCB6 E2BSE3 K7IRE0 V5H2G8 A0A232F303 A0A158NH28 E2AA03 A0A195D1F5 A0A088AK08 I6LNT1 A0A154PPW2 A0A069DR20 A0A0V0G4V8 A0A151XDB4 A0A0P4VL48 R4G3G8 A0A0L7R511 F4WSV6 A0A023F876 A0A0P5A3G8 A0A2R7W062 A0A0P5XMP6 A0A0P4ZI35 A0A1Y1M5A7 A0A182F7Z1 A0A3L8DN99 A0A1L8DLP9 A0A0P5R797 A0A023EKU1 A0A1L8DLZ4 W5JAT2 U5ENM9 A0A310SQ90 Q16JJ2 N6TWA1 A0A0P5BSU5 E9IMS7 A0A182G1Q6 T1E7K4 A0A084W9U3 A0A182XW24 B0XGQ8 A0A0P5XQB8 A0A1A9Z3X0 A0A0L0C1U4 A0A1Q3F5T1 K9IZ15 A0A0M9AA74 A0A1B0D1S2 A0A1B0FNW9 A0A182XGG9 A0A182Q7R6 A0A182N6A4 A0A250YCT2 L5KYI4 A0A182LYA8 A0A2M4AR34 A0A182PNB7 E9GDE7 A0A2C9GRK4 Q7PYU2 A0A1A9VWX7 H2ZZ02 A0A182UY46 A0A1A9XYJ1 A0A3B3QP14 C1BVY9 A0A2K6MEQ0 A0A182LQY2 S7MVZ0 A0A182UAE3 A0A1B0ASR9 A0A1B0CX15 G3TH46 Q7ZWU4 A0A1U7USE1 M3W9V3 A0A2Y9GLN5 A0A2U3XP45 A0A2U3X389 Q3T168 A0A182IN24 A0A182K5T1 A0A2U3V766
Pubmed
19121390
26354079
28756777
22118469
23622113
24845553
+ More
29403074 18362917 19820115 20798317 20075255 28648823 21347285 22539186 26334808 27129103 21719571 25474469 28004739 30249741 24945155 20920257 23761445 17510324 23537049 21282665 26483478 24438588 25244985 26108605 28087693 23258410 21292972 12364791 14747013 17210077 9215903 29240929 20433749 25069045 20966253 27762356 17975172
29403074 18362917 19820115 20798317 20075255 28648823 21347285 22539186 26334808 27129103 21719571 25474469 28004739 30249741 24945155 20920257 23761445 17510324 23537049 21282665 26483478 24438588 25244985 26108605 28087693 23258410 21292972 12364791 14747013 17210077 9215903 29240929 20433749 25069045 20966253 27762356 17975172
EMBL
BABH01012835
KQ459591
KPI97022.1
KQ460597
KPJ13864.1
ODYU01012404
+ More
SOQ58693.1 KZ150082 PZC73840.1 NWSH01006459 PCG63416.1 AGBW02014900 OWR40912.1 GAIX01003316 JAA89244.1 GEBQ01002782 JAT37195.1 GECU01037182 JAS70524.1 NEVH01010537 PNF32263.1 KK852771 KDR16863.1 PYGN01000706 PSN41902.1 GG694600 EFA13571.1 LBMM01021456 KMQ83075.1 GEDC01026194 JAS11104.1 GL450203 EFN81395.1 GALX01001438 JAB67028.1 NNAY01001120 OXU25065.1 ADTU01015494 GL437990 EFN69732.1 KQ976973 KYN06681.1 GU722329 GU722333 KZ288256 ADT65129.1 ADT65133.1 PBC30559.1 KQ434998 KZC13288.1 GBGD01002792 JAC86097.1 GECL01003057 JAP03067.1 KQ982294 KYQ58361.1 GDKW01000758 JAI55837.1 ACPB03007281 GAHY01001691 JAA75819.1 KQ414654 KOC65975.1 GL888327 EGI62763.1 GBBI01001245 JAC17467.1 GDIP01216846 GDIP01216844 JAJ06556.1 KK854210 PTY13133.1 GDIP01082166 JAM21549.1 GDIP01216845 JAJ06557.1 GEZM01040269 JAV80913.1 QOIP01000006 RLU21915.1 GFDF01006698 JAV07386.1 GDIQ01104658 LRGB01000687 JAL47068.1 KZS16746.1 GAPW01003785 JAC09813.1 GFDF01006697 JAV07387.1 ADMH02002023 ETN59945.1 GANO01000529 JAB59342.1 KQ760357 OAD60752.1 CH478006 EAT34449.1 APGK01052695 KB741213 ENN72666.1 GDIP01184854 JAJ38548.1 GL764255 EFZ18148.1 JXUM01038854 KQ561182 KXJ79452.1 GAMD01002957 JAA98633.1 ATLV01021913 KE525325 KFB46987.1 DS233057 EDS27781.1 GDIP01070214 JAM33501.1 JRES01001001 KNC26227.1 GFDL01012217 JAV22828.1 GABZ01007736 JAA45789.1 KQ435715 KOX79113.1 AJVK01010364 CCAG010014421 AXCN02000278 GFFW01003326 JAV41462.1 KB030474 ELK16250.1 AXCM01000395 GGFK01009912 MBW43233.1 GL732540 EFX82491.1 APCN01002014 AAAB01008987 EAA01517.4 AFYH01230726 BT078768 ACO13192.1 KE162497 EPQ08679.1 JXJN01002966 AJWK01033103 BC046706 CM004473 AAH46706.1 OCT82782.1 AANG04001043 BC102091
SOQ58693.1 KZ150082 PZC73840.1 NWSH01006459 PCG63416.1 AGBW02014900 OWR40912.1 GAIX01003316 JAA89244.1 GEBQ01002782 JAT37195.1 GECU01037182 JAS70524.1 NEVH01010537 PNF32263.1 KK852771 KDR16863.1 PYGN01000706 PSN41902.1 GG694600 EFA13571.1 LBMM01021456 KMQ83075.1 GEDC01026194 JAS11104.1 GL450203 EFN81395.1 GALX01001438 JAB67028.1 NNAY01001120 OXU25065.1 ADTU01015494 GL437990 EFN69732.1 KQ976973 KYN06681.1 GU722329 GU722333 KZ288256 ADT65129.1 ADT65133.1 PBC30559.1 KQ434998 KZC13288.1 GBGD01002792 JAC86097.1 GECL01003057 JAP03067.1 KQ982294 KYQ58361.1 GDKW01000758 JAI55837.1 ACPB03007281 GAHY01001691 JAA75819.1 KQ414654 KOC65975.1 GL888327 EGI62763.1 GBBI01001245 JAC17467.1 GDIP01216846 GDIP01216844 JAJ06556.1 KK854210 PTY13133.1 GDIP01082166 JAM21549.1 GDIP01216845 JAJ06557.1 GEZM01040269 JAV80913.1 QOIP01000006 RLU21915.1 GFDF01006698 JAV07386.1 GDIQ01104658 LRGB01000687 JAL47068.1 KZS16746.1 GAPW01003785 JAC09813.1 GFDF01006697 JAV07387.1 ADMH02002023 ETN59945.1 GANO01000529 JAB59342.1 KQ760357 OAD60752.1 CH478006 EAT34449.1 APGK01052695 KB741213 ENN72666.1 GDIP01184854 JAJ38548.1 GL764255 EFZ18148.1 JXUM01038854 KQ561182 KXJ79452.1 GAMD01002957 JAA98633.1 ATLV01021913 KE525325 KFB46987.1 DS233057 EDS27781.1 GDIP01070214 JAM33501.1 JRES01001001 KNC26227.1 GFDL01012217 JAV22828.1 GABZ01007736 JAA45789.1 KQ435715 KOX79113.1 AJVK01010364 CCAG010014421 AXCN02000278 GFFW01003326 JAV41462.1 KB030474 ELK16250.1 AXCM01000395 GGFK01009912 MBW43233.1 GL732540 EFX82491.1 APCN01002014 AAAB01008987 EAA01517.4 AFYH01230726 BT078768 ACO13192.1 KE162497 EPQ08679.1 JXJN01002966 AJWK01033103 BC046706 CM004473 AAH46706.1 OCT82782.1 AANG04001043 BC102091
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000192223
+ More
UP000235965 UP000027135 UP000245037 UP000007266 UP000036403 UP000008237 UP000002358 UP000215335 UP000005205 UP000000311 UP000078542 UP000005203 UP000242457 UP000076502 UP000075809 UP000015103 UP000053825 UP000007755 UP000069272 UP000279307 UP000076858 UP000000673 UP000008820 UP000019118 UP000069940 UP000249989 UP000030765 UP000076408 UP000002320 UP000092445 UP000037069 UP000053105 UP000092462 UP000092444 UP000076407 UP000075886 UP000075884 UP000010552 UP000075883 UP000075885 UP000000305 UP000075840 UP000007062 UP000078200 UP000008672 UP000075903 UP000092443 UP000261540 UP000265140 UP000233180 UP000075882 UP000075902 UP000092460 UP000092461 UP000007646 UP000186698 UP000189704 UP000011712 UP000248481 UP000245341 UP000245340 UP000009136 UP000075880 UP000075881 UP000245320
UP000235965 UP000027135 UP000245037 UP000007266 UP000036403 UP000008237 UP000002358 UP000215335 UP000005205 UP000000311 UP000078542 UP000005203 UP000242457 UP000076502 UP000075809 UP000015103 UP000053825 UP000007755 UP000069272 UP000279307 UP000076858 UP000000673 UP000008820 UP000019118 UP000069940 UP000249989 UP000030765 UP000076408 UP000002320 UP000092445 UP000037069 UP000053105 UP000092462 UP000092444 UP000076407 UP000075886 UP000075884 UP000010552 UP000075883 UP000075885 UP000000305 UP000075840 UP000007062 UP000078200 UP000008672 UP000075903 UP000092443 UP000261540 UP000265140 UP000233180 UP000075882 UP000075902 UP000092460 UP000092461 UP000007646 UP000186698 UP000189704 UP000011712 UP000248481 UP000245341 UP000245340 UP000009136 UP000075880 UP000075881 UP000245320
PRIDE
Pfam
Interpro
IPR007699
SGS_dom
+ More
IPR037893 CS_CacyBP
IPR015120 Siah-Interact_N
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR037201 CacyBP_N
IPR018326 Rad4_beta-hairpin_dom1
IPR036985 Transglutaminase-like_sf
IPR004583 DNA_repair_Rad4
IPR018325 Rad4/PNGase_transGLS-fold
IPR018328 Rad4_beta-hairpin_dom3
IPR018327 BHD_2
IPR038765 Papain-like_cys_pep_sf
IPR037893 CS_CacyBP
IPR015120 Siah-Interact_N
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR037201 CacyBP_N
IPR018326 Rad4_beta-hairpin_dom1
IPR036985 Transglutaminase-like_sf
IPR004583 DNA_repair_Rad4
IPR018325 Rad4/PNGase_transGLS-fold
IPR018328 Rad4_beta-hairpin_dom3
IPR018327 BHD_2
IPR038765 Papain-like_cys_pep_sf
Gene 3D
ProteinModelPortal
H9JHM3
A0A194PUM2
A0A194R8N4
A0A2H1X061
A0A2W1BMA2
A0A2A4IV95
+ More
A0A212EHF9 S4PE51 A0A1B6MMV3 A0A1B6H7D3 A0A1W4WIL7 A0A2J7QUK2 A0A067R418 A0A2P8YCA2 D7GXT5 A0A0J7MRE2 A0A1B6CCB6 E2BSE3 K7IRE0 V5H2G8 A0A232F303 A0A158NH28 E2AA03 A0A195D1F5 A0A088AK08 I6LNT1 A0A154PPW2 A0A069DR20 A0A0V0G4V8 A0A151XDB4 A0A0P4VL48 R4G3G8 A0A0L7R511 F4WSV6 A0A023F876 A0A0P5A3G8 A0A2R7W062 A0A0P5XMP6 A0A0P4ZI35 A0A1Y1M5A7 A0A182F7Z1 A0A3L8DN99 A0A1L8DLP9 A0A0P5R797 A0A023EKU1 A0A1L8DLZ4 W5JAT2 U5ENM9 A0A310SQ90 Q16JJ2 N6TWA1 A0A0P5BSU5 E9IMS7 A0A182G1Q6 T1E7K4 A0A084W9U3 A0A182XW24 B0XGQ8 A0A0P5XQB8 A0A1A9Z3X0 A0A0L0C1U4 A0A1Q3F5T1 K9IZ15 A0A0M9AA74 A0A1B0D1S2 A0A1B0FNW9 A0A182XGG9 A0A182Q7R6 A0A182N6A4 A0A250YCT2 L5KYI4 A0A182LYA8 A0A2M4AR34 A0A182PNB7 E9GDE7 A0A2C9GRK4 Q7PYU2 A0A1A9VWX7 H2ZZ02 A0A182UY46 A0A1A9XYJ1 A0A3B3QP14 C1BVY9 A0A2K6MEQ0 A0A182LQY2 S7MVZ0 A0A182UAE3 A0A1B0ASR9 A0A1B0CX15 G3TH46 Q7ZWU4 A0A1U7USE1 M3W9V3 A0A2Y9GLN5 A0A2U3XP45 A0A2U3X389 Q3T168 A0A182IN24 A0A182K5T1 A0A2U3V766
A0A212EHF9 S4PE51 A0A1B6MMV3 A0A1B6H7D3 A0A1W4WIL7 A0A2J7QUK2 A0A067R418 A0A2P8YCA2 D7GXT5 A0A0J7MRE2 A0A1B6CCB6 E2BSE3 K7IRE0 V5H2G8 A0A232F303 A0A158NH28 E2AA03 A0A195D1F5 A0A088AK08 I6LNT1 A0A154PPW2 A0A069DR20 A0A0V0G4V8 A0A151XDB4 A0A0P4VL48 R4G3G8 A0A0L7R511 F4WSV6 A0A023F876 A0A0P5A3G8 A0A2R7W062 A0A0P5XMP6 A0A0P4ZI35 A0A1Y1M5A7 A0A182F7Z1 A0A3L8DN99 A0A1L8DLP9 A0A0P5R797 A0A023EKU1 A0A1L8DLZ4 W5JAT2 U5ENM9 A0A310SQ90 Q16JJ2 N6TWA1 A0A0P5BSU5 E9IMS7 A0A182G1Q6 T1E7K4 A0A084W9U3 A0A182XW24 B0XGQ8 A0A0P5XQB8 A0A1A9Z3X0 A0A0L0C1U4 A0A1Q3F5T1 K9IZ15 A0A0M9AA74 A0A1B0D1S2 A0A1B0FNW9 A0A182XGG9 A0A182Q7R6 A0A182N6A4 A0A250YCT2 L5KYI4 A0A182LYA8 A0A2M4AR34 A0A182PNB7 E9GDE7 A0A2C9GRK4 Q7PYU2 A0A1A9VWX7 H2ZZ02 A0A182UY46 A0A1A9XYJ1 A0A3B3QP14 C1BVY9 A0A2K6MEQ0 A0A182LQY2 S7MVZ0 A0A182UAE3 A0A1B0ASR9 A0A1B0CX15 G3TH46 Q7ZWU4 A0A1U7USE1 M3W9V3 A0A2Y9GLN5 A0A2U3XP45 A0A2U3X389 Q3T168 A0A182IN24 A0A182K5T1 A0A2U3V766
PDB
1X5M
E-value=1.55119e-22,
Score=259
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
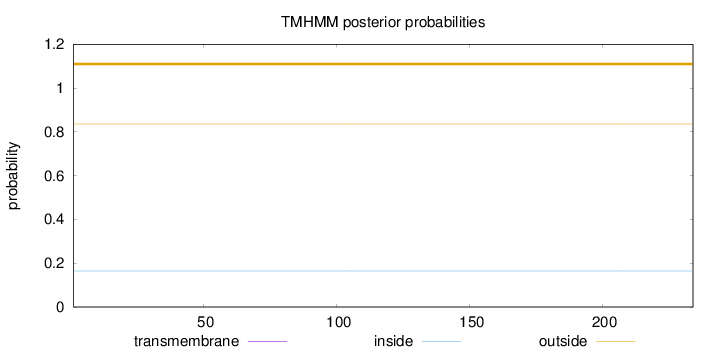
Length:
234
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.16410
outside
1 - 234
Population Genetic Test Statistics
Pi
169.626629
Theta
142.623585
Tajima's D
0.595008
CLR
0.902919
CSRT
0.544522773861307
Interpretation
Uncertain
