Pre Gene Modal
BGIBMGA009022
Annotation
Group_XIIA_secretory_phospholipase_A2_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 3.124
Sequence
CDS
ATGTTTCGTAATGTTATTACTGTTGCCGAGAAATTTAAATCTTTACATGATGTATTCGATGCAGCCGTAGAAGAAGAATGTATATTTACTTGTCCAGGAGGCCAAAAGCCAGTTAGAAACCGAAATCATATACCGAAATCTGATGGATGTGGTTCATTGGGATTTGAAATATCATCGGAATACTTACCATTAAATGAGATGACAAAATGCTGTGATGCTCATGATATTTGTTACGACACATGCAACAGCGGAAAAGAAGTATGTGACCTAGAGTTTAAGAGATGTCTCTATAATTACTGCAATTCATATAAATCTGTGAACATTGCTGGTGAGACTATCACTAAAGGTTGTAAAGGTGCAGCTAAATTGCTATTTACAGGAACACTTACATTAGGATGTAAATCATATTTGGATGCACAGAAGAATGCTTGTTACTGTCCAGACAAGAATACCAAGTACAAGAAATATTCAGGAAATGGTGAATTGTAG
Protein
MFRNVITVAEKFKSLHDVFDAAVEEECIFTCPGGQKPVRNRNHIPKSDGCGSLGFEISSEYLPLNEMTKCCDAHDICYDTCNSGKEVCDLEFKRCLYNYCNSYKSVNIAGETITKGCKGAAKLLFTGTLTLGCKSYLDAQKNACYCPDKNTKYKKYSGNGEL
Summary
Similarity
Belongs to the phospholipase A2 family.
Uniprot
H9JHM4
A0A194PW88
A0A194RD80
A0A2W1BQ63
A0A2H1WAD7
A0A2A4IWB1
+ More
A0A212EHE8 S4PIT2 A0A0L7LDW5 A0A1W4WIS5 D6WWD0 A0A195DP69 A0A151WZ86 A0A2M4BYN2 T1DIQ5 A0A0N0BJJ2 W5JTT2 A0A2M4BZF5 T1EBA3 A0A2M3ZD22 A0A1J1IMZ4 A0A182FW24 A0A2M4AQN7 A0A195BHB2 A0A158P350 A0A195CD99 F4W4H5 A0A182R6M5 Q7QHA9 A0A195EZK9 A0A182YAX3 A0A182M743 A0A182P5E9 A0A026W3P0 A0A182JP15 A0A310SU71 A0A154PIV1 A0A182WDP5 A0A182N256 A0A182QCR6 T1DER7 A0A1B0B1D7 A0A1A9XSS5 A0A1Y1L8A5 A0A2A3EN98 A0A023EKT1 A0A087ZS81 Q16HL6 U5EF81 A0A084W8B7 E9IIK4 A0A1A9VHN5 A0A1B0D6Y2 A0A0J7NQD7 A0A1B0AFZ2 D3TQ71 B0WPR5 B4J3Q0 A0A0L7RCQ1 B4KYJ0 A0A1Q3FJT5 B3M9V1 B4LDY6 J3JTY7 A0A182ILK2 B4MXV2 A0A1I8PBV4 E2A1C1 K7IPQ1 A0A1I8NDB0 A0A1A9W505 N6T0R9 A0A067RM52 A0A0M4EIY7 B4ITX1 B3NDP0 B4HIK8 B4QLR1 Q9VUV6 A0A0J9ULZ7 A0A3B0KH08 A0A336M9P2 Q2M0J2 B4GRJ9 A0A1W4VB56 A0A182HW65 A0A182KKX2 A0A182UBU7 W8BPI3 A0A0K8UYH7 A0A182X5T0 A0A034WGY9 A0A0A1XQC9 A0A336MDW0 A0A2M4AVQ6 A0A1B0GJ13 A0A2Z4BXT7 A0A0L0C107 A0A182VMU7 A0A1B6GL72
A0A212EHE8 S4PIT2 A0A0L7LDW5 A0A1W4WIS5 D6WWD0 A0A195DP69 A0A151WZ86 A0A2M4BYN2 T1DIQ5 A0A0N0BJJ2 W5JTT2 A0A2M4BZF5 T1EBA3 A0A2M3ZD22 A0A1J1IMZ4 A0A182FW24 A0A2M4AQN7 A0A195BHB2 A0A158P350 A0A195CD99 F4W4H5 A0A182R6M5 Q7QHA9 A0A195EZK9 A0A182YAX3 A0A182M743 A0A182P5E9 A0A026W3P0 A0A182JP15 A0A310SU71 A0A154PIV1 A0A182WDP5 A0A182N256 A0A182QCR6 T1DER7 A0A1B0B1D7 A0A1A9XSS5 A0A1Y1L8A5 A0A2A3EN98 A0A023EKT1 A0A087ZS81 Q16HL6 U5EF81 A0A084W8B7 E9IIK4 A0A1A9VHN5 A0A1B0D6Y2 A0A0J7NQD7 A0A1B0AFZ2 D3TQ71 B0WPR5 B4J3Q0 A0A0L7RCQ1 B4KYJ0 A0A1Q3FJT5 B3M9V1 B4LDY6 J3JTY7 A0A182ILK2 B4MXV2 A0A1I8PBV4 E2A1C1 K7IPQ1 A0A1I8NDB0 A0A1A9W505 N6T0R9 A0A067RM52 A0A0M4EIY7 B4ITX1 B3NDP0 B4HIK8 B4QLR1 Q9VUV6 A0A0J9ULZ7 A0A3B0KH08 A0A336M9P2 Q2M0J2 B4GRJ9 A0A1W4VB56 A0A182HW65 A0A182KKX2 A0A182UBU7 W8BPI3 A0A0K8UYH7 A0A182X5T0 A0A034WGY9 A0A0A1XQC9 A0A336MDW0 A0A2M4AVQ6 A0A1B0GJ13 A0A2Z4BXT7 A0A0L0C107 A0A182VMU7 A0A1B6GL72
Pubmed
19121390
26354079
28756777
22118469
23622113
26227816
+ More
18362917 19820115 20920257 23761445 21347285 21719571 12364791 14747013 17210077 25244985 24508170 30249741 24330624 28004739 24945155 26483478 17510324 24438588 21282665 20353571 17994087 22516182 23537049 20798317 20075255 25315136 24845553 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20966253 24495485 25348373 25830018 26108605
18362917 19820115 20920257 23761445 21347285 21719571 12364791 14747013 17210077 25244985 24508170 30249741 24330624 28004739 24945155 26483478 17510324 24438588 21282665 20353571 17994087 22516182 23537049 20798317 20075255 25315136 24845553 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20966253 24495485 25348373 25830018 26108605
EMBL
BABH01012837
KQ459591
KPI97024.1
KQ460597
KPJ13866.1
KZ150082
+ More
PZC73843.1 ODYU01007223 SOQ49812.1 NWSH01006459 PCG63420.1 AGBW02014900 OWR40909.1 GAIX01000079 JAA92481.1 JTDY01001603 KOB73396.1 KQ971361 EFA08691.1 KQ980713 KYN14284.1 KQ982649 KYQ53077.1 GGFJ01009056 MBW58197.1 GAMD01001811 JAA99779.1 KQ435714 KOX79218.1 ADMH02000399 ETN66693.1 GGFJ01009292 MBW58433.1 GAMD01000048 JAB01543.1 GGFM01005627 MBW26378.1 CVRI01000055 CRL01605.1 GGFK01009762 MBW43083.1 KQ976467 KYM84159.1 ADTU01007751 KQ977935 KYM98685.1 GL887532 EGI70870.1 AAAB01008816 EAA05247.3 KQ981905 KYN33561.1 AXCM01001638 KK107447 QOIP01000001 EZA50705.1 RLU26341.1 KQ760382 OAD60693.1 KQ434931 KZC11779.1 AXCN02001743 GALA01000932 JAA93920.1 JXJN01007077 GEZM01063663 JAV69031.1 KZ288204 PBC33208.1 JXUM01123682 GAPW01003807 KQ566744 JAC09791.1 KXJ69857.1 CH478160 EAT33743.1 EAT33745.1 EJY58073.1 GANO01003979 JAB55892.1 ATLV01021410 KE525318 KFB46461.1 GL763494 EFZ19572.1 AJVK01003776 LBMM01002569 KMQ94650.1 CCAG010010400 EZ423573 ADD19849.1 DS232028 EDS32474.1 CH916366 EDV97281.1 KQ414615 KOC68767.1 CH933809 EDW18801.1 GFDL01007292 JAV27753.1 CH902618 EDV40142.1 CH940647 EDW70029.1 BT126697 KB631611 AEE61659.1 ERL84648.1 CH963876 EDW76871.1 GL435760 EFN72747.1 APGK01056605 APGK01056606 KB741277 ENN71078.1 KK852544 KDR21645.1 CP012525 ALC44802.1 CH891734 EDW99834.1 CH954178 EDV52173.1 CH480815 EDW41638.1 CM000363 CM002912 EDX10610.1 KMY99879.1 AE014296 AF349541 AAF49567.2 AAK30169.1 AGB94596.1 KMY99880.1 OUUW01000009 SPP85016.1 UFQT01000755 SSX26996.1 CH379069 EAL30940.2 CH479188 EDW40384.1 APCN01001469 GAMC01005718 JAC00838.1 GDHF01020711 JAI31603.1 GAKP01005547 JAC53405.1 GBXI01005675 GBXI01001130 JAD08617.1 JAD13162.1 UFQT01000940 SSX28120.1 GGFK01011387 MBW44708.1 AJWK01019202 AJWK01019203 AJWK01019204 MH055790 AWU67138.1 JRES01001048 KNC25975.1 GECZ01006628 JAS63141.1
PZC73843.1 ODYU01007223 SOQ49812.1 NWSH01006459 PCG63420.1 AGBW02014900 OWR40909.1 GAIX01000079 JAA92481.1 JTDY01001603 KOB73396.1 KQ971361 EFA08691.1 KQ980713 KYN14284.1 KQ982649 KYQ53077.1 GGFJ01009056 MBW58197.1 GAMD01001811 JAA99779.1 KQ435714 KOX79218.1 ADMH02000399 ETN66693.1 GGFJ01009292 MBW58433.1 GAMD01000048 JAB01543.1 GGFM01005627 MBW26378.1 CVRI01000055 CRL01605.1 GGFK01009762 MBW43083.1 KQ976467 KYM84159.1 ADTU01007751 KQ977935 KYM98685.1 GL887532 EGI70870.1 AAAB01008816 EAA05247.3 KQ981905 KYN33561.1 AXCM01001638 KK107447 QOIP01000001 EZA50705.1 RLU26341.1 KQ760382 OAD60693.1 KQ434931 KZC11779.1 AXCN02001743 GALA01000932 JAA93920.1 JXJN01007077 GEZM01063663 JAV69031.1 KZ288204 PBC33208.1 JXUM01123682 GAPW01003807 KQ566744 JAC09791.1 KXJ69857.1 CH478160 EAT33743.1 EAT33745.1 EJY58073.1 GANO01003979 JAB55892.1 ATLV01021410 KE525318 KFB46461.1 GL763494 EFZ19572.1 AJVK01003776 LBMM01002569 KMQ94650.1 CCAG010010400 EZ423573 ADD19849.1 DS232028 EDS32474.1 CH916366 EDV97281.1 KQ414615 KOC68767.1 CH933809 EDW18801.1 GFDL01007292 JAV27753.1 CH902618 EDV40142.1 CH940647 EDW70029.1 BT126697 KB631611 AEE61659.1 ERL84648.1 CH963876 EDW76871.1 GL435760 EFN72747.1 APGK01056605 APGK01056606 KB741277 ENN71078.1 KK852544 KDR21645.1 CP012525 ALC44802.1 CH891734 EDW99834.1 CH954178 EDV52173.1 CH480815 EDW41638.1 CM000363 CM002912 EDX10610.1 KMY99879.1 AE014296 AF349541 AAF49567.2 AAK30169.1 AGB94596.1 KMY99880.1 OUUW01000009 SPP85016.1 UFQT01000755 SSX26996.1 CH379069 EAL30940.2 CH479188 EDW40384.1 APCN01001469 GAMC01005718 JAC00838.1 GDHF01020711 JAI31603.1 GAKP01005547 JAC53405.1 GBXI01005675 GBXI01001130 JAD08617.1 JAD13162.1 UFQT01000940 SSX28120.1 GGFK01011387 MBW44708.1 AJWK01019202 AJWK01019203 AJWK01019204 MH055790 AWU67138.1 JRES01001048 KNC25975.1 GECZ01006628 JAS63141.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000192223 UP000007266 UP000078492 UP000075809 UP000053105 UP000000673 UP000183832 UP000069272 UP000078540 UP000005205 UP000078542 UP000007755 UP000075900 UP000007062 UP000078541 UP000076408 UP000075883 UP000075885 UP000053097 UP000279307 UP000075881 UP000076502 UP000075920 UP000075884 UP000075886 UP000092460 UP000092443 UP000242457 UP000069940 UP000249989 UP000005203 UP000008820 UP000030765 UP000078200 UP000092462 UP000036403 UP000092445 UP000092444 UP000002320 UP000001070 UP000053825 UP000009192 UP000007801 UP000008792 UP000030742 UP000075880 UP000007798 UP000095300 UP000000311 UP000002358 UP000095301 UP000091820 UP000019118 UP000027135 UP000092553 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000268350 UP000001819 UP000008744 UP000192221 UP000075840 UP000075882 UP000075902 UP000076407 UP000092461 UP000037069 UP000075903
UP000192223 UP000007266 UP000078492 UP000075809 UP000053105 UP000000673 UP000183832 UP000069272 UP000078540 UP000005205 UP000078542 UP000007755 UP000075900 UP000007062 UP000078541 UP000076408 UP000075883 UP000075885 UP000053097 UP000279307 UP000075881 UP000076502 UP000075920 UP000075884 UP000075886 UP000092460 UP000092443 UP000242457 UP000069940 UP000249989 UP000005203 UP000008820 UP000030765 UP000078200 UP000092462 UP000036403 UP000092445 UP000092444 UP000002320 UP000001070 UP000053825 UP000009192 UP000007801 UP000008792 UP000030742 UP000075880 UP000007798 UP000095300 UP000000311 UP000002358 UP000095301 UP000091820 UP000019118 UP000027135 UP000092553 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000268350 UP000001819 UP000008744 UP000192221 UP000075840 UP000075882 UP000075902 UP000076407 UP000092461 UP000037069 UP000075903
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JHM4
A0A194PW88
A0A194RD80
A0A2W1BQ63
A0A2H1WAD7
A0A2A4IWB1
+ More
A0A212EHE8 S4PIT2 A0A0L7LDW5 A0A1W4WIS5 D6WWD0 A0A195DP69 A0A151WZ86 A0A2M4BYN2 T1DIQ5 A0A0N0BJJ2 W5JTT2 A0A2M4BZF5 T1EBA3 A0A2M3ZD22 A0A1J1IMZ4 A0A182FW24 A0A2M4AQN7 A0A195BHB2 A0A158P350 A0A195CD99 F4W4H5 A0A182R6M5 Q7QHA9 A0A195EZK9 A0A182YAX3 A0A182M743 A0A182P5E9 A0A026W3P0 A0A182JP15 A0A310SU71 A0A154PIV1 A0A182WDP5 A0A182N256 A0A182QCR6 T1DER7 A0A1B0B1D7 A0A1A9XSS5 A0A1Y1L8A5 A0A2A3EN98 A0A023EKT1 A0A087ZS81 Q16HL6 U5EF81 A0A084W8B7 E9IIK4 A0A1A9VHN5 A0A1B0D6Y2 A0A0J7NQD7 A0A1B0AFZ2 D3TQ71 B0WPR5 B4J3Q0 A0A0L7RCQ1 B4KYJ0 A0A1Q3FJT5 B3M9V1 B4LDY6 J3JTY7 A0A182ILK2 B4MXV2 A0A1I8PBV4 E2A1C1 K7IPQ1 A0A1I8NDB0 A0A1A9W505 N6T0R9 A0A067RM52 A0A0M4EIY7 B4ITX1 B3NDP0 B4HIK8 B4QLR1 Q9VUV6 A0A0J9ULZ7 A0A3B0KH08 A0A336M9P2 Q2M0J2 B4GRJ9 A0A1W4VB56 A0A182HW65 A0A182KKX2 A0A182UBU7 W8BPI3 A0A0K8UYH7 A0A182X5T0 A0A034WGY9 A0A0A1XQC9 A0A336MDW0 A0A2M4AVQ6 A0A1B0GJ13 A0A2Z4BXT7 A0A0L0C107 A0A182VMU7 A0A1B6GL72
A0A212EHE8 S4PIT2 A0A0L7LDW5 A0A1W4WIS5 D6WWD0 A0A195DP69 A0A151WZ86 A0A2M4BYN2 T1DIQ5 A0A0N0BJJ2 W5JTT2 A0A2M4BZF5 T1EBA3 A0A2M3ZD22 A0A1J1IMZ4 A0A182FW24 A0A2M4AQN7 A0A195BHB2 A0A158P350 A0A195CD99 F4W4H5 A0A182R6M5 Q7QHA9 A0A195EZK9 A0A182YAX3 A0A182M743 A0A182P5E9 A0A026W3P0 A0A182JP15 A0A310SU71 A0A154PIV1 A0A182WDP5 A0A182N256 A0A182QCR6 T1DER7 A0A1B0B1D7 A0A1A9XSS5 A0A1Y1L8A5 A0A2A3EN98 A0A023EKT1 A0A087ZS81 Q16HL6 U5EF81 A0A084W8B7 E9IIK4 A0A1A9VHN5 A0A1B0D6Y2 A0A0J7NQD7 A0A1B0AFZ2 D3TQ71 B0WPR5 B4J3Q0 A0A0L7RCQ1 B4KYJ0 A0A1Q3FJT5 B3M9V1 B4LDY6 J3JTY7 A0A182ILK2 B4MXV2 A0A1I8PBV4 E2A1C1 K7IPQ1 A0A1I8NDB0 A0A1A9W505 N6T0R9 A0A067RM52 A0A0M4EIY7 B4ITX1 B3NDP0 B4HIK8 B4QLR1 Q9VUV6 A0A0J9ULZ7 A0A3B0KH08 A0A336M9P2 Q2M0J2 B4GRJ9 A0A1W4VB56 A0A182HW65 A0A182KKX2 A0A182UBU7 W8BPI3 A0A0K8UYH7 A0A182X5T0 A0A034WGY9 A0A0A1XQC9 A0A336MDW0 A0A2M4AVQ6 A0A1B0GJ13 A0A2Z4BXT7 A0A0L0C107 A0A182VMU7 A0A1B6GL72
Ontologies
PATHWAY
00564
Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
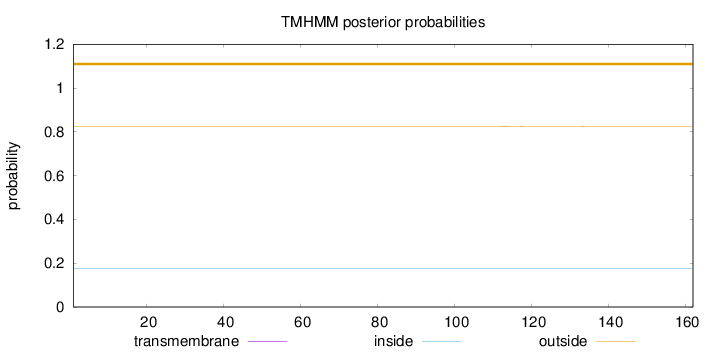
Topology
Subcellular location
Secreted
Length:
162
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0049
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17484
outside
1 - 162
Population Genetic Test Statistics
Pi
206.532101
Theta
150.394367
Tajima's D
1.173407
CLR
0
CSRT
0.706214689265537
Interpretation
Uncertain
