Gene
KWMTBOMO01263 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009027
Annotation
B_Chain_B?_Crystal_Structure_Of_Bombyx_Mori_Sp2/sp3_Heterohexamer
Full name
Arylphorin subunit alpha
+ More
Arylphorin subunit beta
Arylphorin
Sex-specific storage-protein 2
Arylphorin subunit beta
Arylphorin
Sex-specific storage-protein 2
Alternative Name
Arylphorin
Location in the cell
Extracellular Reliability : 3.144
Sequence
CDS
ATGAAGACTGTCTTGATTTTAGCGGGGCTTATTGCCCTCGCTCTCAGTAGCACCGTACCTGAATTCAAAACGACACCGGTTGACGCTGCGTTCGTCGAAAAGCAAAAGAAGATTTTGTCTCTTTTCTACAACGTTAACGAGATCAGCTATGAAGCTGAGTACTACAAAGTCGCCCAGGACTTCAACATCGAGGCCAGCAAGGACTGCTACACAAACATGAAAGCCTACGAAAATTTCATGATGATGTACAAGGTCGGATTCCTTCCCAAGAATTTGGAATTCTCGATCTTTTACGAAAAAATGAGGGAAGAAGCCATCGCGCTGTTCAAGCTGTTCTATTATGCCAAAGATTTTGAATGTTTCTACAAAACAGCATGCTACGCCAGAGTCTACATGAACCAGGGAATGTTCTTATACGCCTACTACATAGCTATTATCCAGCGCTCTGACACCGCCAGCTTCGTTCTACCTGCTCCATACGAAGCTTATCCTCAATATTTTGTCAACATGGAAGTTAAAAATAAAATGGACTACGTTAAGATGATGGATGGTTGCCTTGACGAGAAAATATGTTATAATTACGGAATTATCAAAGAAAACGAACAATTCGTAATGTACGCCAACTATTCCAATTCCCTGACTTACCCCAACAACGAAGACAGAATTGCTTACTTAACTGAAGATGTTGGCCTAAATGCTTATTATTACTATTTCCACTCACACTTACCGTTCTGGTGGAACTCTGGTAAATACGGAGCTTTCAAGGAACGTCGTGGGGAAATCTACTTCTTCTTCTACCAACAACTGTTAGCTCGTTACTACATGGAGCGTCTTACCAATGGATTGGGTAAAATCCCTGAGTTTTCCTGGTACTCGCCTCTTCGAACTGGCTACCTCCCGCCTTTCAACTCCTTTTATTATCCATTTGCCCAAAGAAGCAATGACTACGAGTTGCACACCGAGAAGAACTACGAAGAAATTCGCTTCCTCGACATTTATGAGAAGACATTCTTCCAATACCTCCAGCAAGGTCACTTCAAGGCCTTCGACAAAAAAATTGATCTGCACAGTAGCAAAGCAGTCAACTTTGTGGGCAACTACTGGCAGACGAACGCCGATCTATTCGAAGAAGACTTCCTCCAATTCTACCAGCGTTCTTACGAGGTTAACGCTCGCCGCGTACTTGGTGCTGCTCCTAAGCCTTTCAACCAGTACACATTTATTCCTAGTGCTCTTGACTTCTACCAAACTTCGGCCAGGGACCCAGCCTTCTACCAGCTCTATAAAAGAATTGTTCAATACATCATCGAATTTAAACAGTACCAGGTACCATACACACAAGAAGCTCTTCACTTCGTTGGTCTGAAAATTTCCGACGTCAAAGTCGACAAAATGGTCACATTCTTTGACCATTTCGACTTCGATGCCTTCAACACTGTCTACTTCAGTAAAGAAGAACTCAAGAGCTCTCCTCACGGTTACAAGGTCCGTCAACCACGTCTTAACCACAAGCCGTTCACTGTGACAATCGATATTAAATCTGATGTTGCCACTAACGCCGTCGTCAAGATGTTCCTTGGACCTAAATACGATGAGAACGGCTTTCCCTTCAGCCTAGAAGATAACTGGATGAACTTCTACGAGCTTGACTGGTTCGTGCAAAAGGTTAACCCTGGCCAGTCTCAGATAACGCGTTCATCTACTGACTTTGCATTCTTCAAAGAAGACTCTTTACCGATGGCCGAAATCTACAAGCTCTTGGACCAAGGAAAGATTCCTACTGACATGTTCAACTCCTCGGACACTATGCCTTCGAGGCTGATGCTGCCTAAGGGTACATACGATGGATTCCCCTTCCAGCTGTTTGTATTCGTCTATCCGTATGAACCGACACCTAAGGAGTCAGAGCCATTCAAGTCTGTTGTTCCGGACAACAAACCATTCGGTTATCCATTCGATCGCCCCGTTCTTCCTCAGTACTTCAAACAACCTAACATGTTCTTCAAGAAGGTCCTGGTCTACCATGAAGGAGAACTATTCCCCTATCTATTTAACATTCCTCACTATACACCAGATAAAGCGCAACTATAA
Protein
MKTVLILAGLIALALSSTVPEFKTTPVDAAFVEKQKKILSLFYNVNEISYEAEYYKVAQDFNIEASKDCYTNMKAYENFMMMYKVGFLPKNLEFSIFYEKMREEAIALFKLFYYAKDFECFYKTACYARVYMNQGMFLYAYYIAIIQRSDTASFVLPAPYEAYPQYFVNMEVKNKMDYVKMMDGCLDEKICYNYGIIKENEQFVMYANYSNSLTYPNNEDRIAYLTEDVGLNAYYYYFHSHLPFWWNSGKYGAFKERRGEIYFFFYQQLLARYYMERLTNGLGKIPEFSWYSPLRTGYLPPFNSFYYPFAQRSNDYELHTEKNYEEIRFLDIYEKTFFQYLQQGHFKAFDKKIDLHSSKAVNFVGNYWQTNADLFEEDFLQFYQRSYEVNARRVLGAAPKPFNQYTFIPSALDFYQTSARDPAFYQLYKRIVQYIIEFKQYQVPYTQEALHFVGLKISDVKVDKMVTFFDHFDFDAFNTVYFSKEELKSSPHGYKVRQPRLNHKPFTVTIDIKSDVATNAVVKMFLGPKYDENGFPFSLEDNWMNFYELDWFVQKVNPGQSQITRSSTDFAFFKEDSLPMAEIYKLLDQGKIPTDMFNSSDTMPSRLMLPKGTYDGFPFQLFVFVYPYEPTPKESEPFKSVVPDNKPFGYPFDRPVLPQYFKQPNMFFKKVLVYHEGELFPYLFNIPHYTPDKAQL
Summary
Description
Arylphorin is a larval storage protein (LSP) which may serve as a storage protein used primarily as a source of aromatic amino acids for protein synthesis during metamorphosis. It is a constituent of the sclerotizing system of the cuticle, and serves as a carrier for ecdysteroid hormone (By similarity).
Subunit
Arylphorin is a hexamer of subunits alpha and beta.
Homohexamer of two stacked trimers; disulfide-linked.
Homohexamer of two stacked trimers; disulfide-linked.
Similarity
Belongs to the hemocyanin family.
Keywords
Glycoprotein
Secreted
Signal
Storage protein
3D-structure
Disulfide bond
Complete proteome
Reference proteome
Feature
chain Arylphorin subunit alpha
Uniprot
H9JHM9
G9I6Y1
P14296
Q1HPP4
A1ILK0
O17478
+ More
P14297 U3KV76 A0A2W1BLW4 U3KV63 Q7Z1F8 A0A3G1NIA0 A0A2W1BJI5 P20613 A0A1B3PEI2 G3LF43 A0A2H1WEX3 A0A2A4J4V2 A0A2A4IW12 A0A0L7LM90 A0A3G1NI73 A0A194R7R7 I4DIH8 A0A194PW98 Q9U5K4 A0A1E1W5U0 A0A212ENI8 A0A1E1WLP6 A0A2H1WFB5 Q24995 A0A1E1WJ47 C6ETM8 Q9GQ56 Q9GQ55 A0A088G3X5 Q0EAG4 D3XLC1 A0A1Z2RRG4 Q0WYG7 Q0EAG5 A4GTP1 Q16XN3 A0A182GR26 B0WKQ2 A0A182F863 A0A182FQC3 W5J942 Q16I89 W5JSY3 Q16QU8 A0A1S4FSI4 A0A182H457 T1DNH0 B0W460 A0A182H055 A0A182Q072 A0A182MT47 Q7PUR0 A0A182X9P1 A0A182TGU7 A0A182L8X1 A0A182HKM5 A0A182R4M0 T1EA57 A0A182UUP7 A0A182Y3M6 P90664 A0A182PGJ5 A0A182NS01 A0A182VLH6 A0A182RZ96 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182WSL9 A0A182VZG2 Q7PXY7 A0A182JQ42 A0A182YGB4 A0A182VR52 A0A182MBJ5 A0A182J7X8 A0A182IRZ0 A0A182P2L1 A0A182KE62 A0A182S9W9 A0A084VS76 B0W462 A0A1B0D919 A0A1B0GLC3 A0A182Q5V2 A0A182HAP9 Q16I87 A0A1S4FZY0 A0A1J1J6D1 A0A182NSB7 A0A182IZN6 A0A084WBS9 A0A182QUT9
P14297 U3KV76 A0A2W1BLW4 U3KV63 Q7Z1F8 A0A3G1NIA0 A0A2W1BJI5 P20613 A0A1B3PEI2 G3LF43 A0A2H1WEX3 A0A2A4J4V2 A0A2A4IW12 A0A0L7LM90 A0A3G1NI73 A0A194R7R7 I4DIH8 A0A194PW98 Q9U5K4 A0A1E1W5U0 A0A212ENI8 A0A1E1WLP6 A0A2H1WFB5 Q24995 A0A1E1WJ47 C6ETM8 Q9GQ56 Q9GQ55 A0A088G3X5 Q0EAG4 D3XLC1 A0A1Z2RRG4 Q0WYG7 Q0EAG5 A4GTP1 Q16XN3 A0A182GR26 B0WKQ2 A0A182F863 A0A182FQC3 W5J942 Q16I89 W5JSY3 Q16QU8 A0A1S4FSI4 A0A182H457 T1DNH0 B0W460 A0A182H055 A0A182Q072 A0A182MT47 Q7PUR0 A0A182X9P1 A0A182TGU7 A0A182L8X1 A0A182HKM5 A0A182R4M0 T1EA57 A0A182UUP7 A0A182Y3M6 P90664 A0A182PGJ5 A0A182NS01 A0A182VLH6 A0A182RZ96 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182WSL9 A0A182VZG2 Q7PXY7 A0A182JQ42 A0A182YGB4 A0A182VR52 A0A182MBJ5 A0A182J7X8 A0A182IRZ0 A0A182P2L1 A0A182KE62 A0A182S9W9 A0A084VS76 B0W462 A0A1B0D919 A0A1B0GLC3 A0A182Q5V2 A0A182HAP9 Q16I87 A0A1S4FZY0 A0A1J1J6D1 A0A182NSB7 A0A182IZN6 A0A084WBS9 A0A182QUT9
Pubmed
EMBL
BABH01012849
JQ043182
AFD02109.1
JN581664
AET36899.1
M28394
+ More
M28396 BABH01012853 DQ443358 ABF51447.1 AB288052 BAF42699.1 AF032396 AAB86644.1 M28395 M28397 JN676156 AFP55241.1 KZ150082 PZC73856.1 JN676155 AFP55240.1 AY278025 EF597564 D44483 KY419217 AVC68643.1 PZC73855.1 M24370 M24371 AB019209 KX462992 AOG20786.1 JN315688 AEO51737.1 ODYU01008226 SOQ51635.1 NWSH01003382 PCG66422.1 NWSH01005808 PCG63931.1 JTDY01000570 KOB76643.1 KY419214 AVC68640.1 KQ460597 KPJ13878.1 AK401096 BAM17718.1 KQ459591 KPI97034.1 AJ249471 CAB55605.1 GDQN01008711 JAT82343.1 AGBW02013638 OWR43054.1 GDQN01003207 JAT87847.1 SOQ51636.1 M73793 AAA74229.1 GDQN01004040 JAT87014.1 EU661547 ACF70482.1 AF294808 AAG44959.1 AF294809 AAG44960.1 KF687948 AIM56832.1 AB253735 BAF32562.1 GU362706 ADC68623.1 KY924788 ASA46442.1 AB248058 BAE98325.1 AB253734 BAF32561.1 EF429084 ABO27097.1 CH477534 EAT39409.1 JXUM01081717 KQ563264 KXJ74179.1 DS231974 EDS29983.1 ADMH02002026 ETN59893.1 CH478096 EAT33977.1 ADMH02000246 ETN67251.1 CH477731 EAT36773.1 JXUM01025392 KQ560720 KXJ81155.1 GAMD01002885 JAA98705.1 DS231835 EDS32904.1 JXUM01100592 KQ564582 KXJ72065.1 AXCN02000261 AXCM01007233 AAAB01008987 EAA01166.5 APCN01000846 GAMD01000727 JAB00864.1 U86080 AAB46714.1 APCN01000900 EAA01337.5 AXCM01011608 ATLV01015833 KE525036 KFB40820.1 EDS32906.1 AJVK01012873 AJWK01034743 AXCN02000206 JXUM01123104 KQ566681 KXJ69911.1 EAT33979.1 CVRI01000070 CRL07340.1 ATLV01022440 KE525332 KFB47673.1
M28396 BABH01012853 DQ443358 ABF51447.1 AB288052 BAF42699.1 AF032396 AAB86644.1 M28395 M28397 JN676156 AFP55241.1 KZ150082 PZC73856.1 JN676155 AFP55240.1 AY278025 EF597564 D44483 KY419217 AVC68643.1 PZC73855.1 M24370 M24371 AB019209 KX462992 AOG20786.1 JN315688 AEO51737.1 ODYU01008226 SOQ51635.1 NWSH01003382 PCG66422.1 NWSH01005808 PCG63931.1 JTDY01000570 KOB76643.1 KY419214 AVC68640.1 KQ460597 KPJ13878.1 AK401096 BAM17718.1 KQ459591 KPI97034.1 AJ249471 CAB55605.1 GDQN01008711 JAT82343.1 AGBW02013638 OWR43054.1 GDQN01003207 JAT87847.1 SOQ51636.1 M73793 AAA74229.1 GDQN01004040 JAT87014.1 EU661547 ACF70482.1 AF294808 AAG44959.1 AF294809 AAG44960.1 KF687948 AIM56832.1 AB253735 BAF32562.1 GU362706 ADC68623.1 KY924788 ASA46442.1 AB248058 BAE98325.1 AB253734 BAF32561.1 EF429084 ABO27097.1 CH477534 EAT39409.1 JXUM01081717 KQ563264 KXJ74179.1 DS231974 EDS29983.1 ADMH02002026 ETN59893.1 CH478096 EAT33977.1 ADMH02000246 ETN67251.1 CH477731 EAT36773.1 JXUM01025392 KQ560720 KXJ81155.1 GAMD01002885 JAA98705.1 DS231835 EDS32904.1 JXUM01100592 KQ564582 KXJ72065.1 AXCN02000261 AXCM01007233 AAAB01008987 EAA01166.5 APCN01000846 GAMD01000727 JAB00864.1 U86080 AAB46714.1 APCN01000900 EAA01337.5 AXCM01011608 ATLV01015833 KE525036 KFB40820.1 EDS32906.1 AJVK01012873 AJWK01034743 AXCN02000206 JXUM01123104 KQ566681 KXJ69911.1 EAT33979.1 CVRI01000070 CRL07340.1 ATLV01022440 KE525332 KFB47673.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000069272 UP000000673 UP000075886 UP000075883 UP000007062 UP000076407 UP000075902 UP000075882 UP000075840 UP000075900 UP000075903 UP000076408 UP000075885 UP000075884 UP000075920 UP000075881 UP000075880 UP000075901 UP000030765 UP000092462 UP000092461 UP000183832
UP000008820 UP000069940 UP000249989 UP000002320 UP000069272 UP000000673 UP000075886 UP000075883 UP000007062 UP000076407 UP000075902 UP000075882 UP000075840 UP000075900 UP000075903 UP000076408 UP000075885 UP000075884 UP000075920 UP000075881 UP000075880 UP000075901 UP000030765 UP000092462 UP000092461 UP000183832
Interpro
Gene 3D
ProteinModelPortal
H9JHM9
G9I6Y1
P14296
Q1HPP4
A1ILK0
O17478
+ More
P14297 U3KV76 A0A2W1BLW4 U3KV63 Q7Z1F8 A0A3G1NIA0 A0A2W1BJI5 P20613 A0A1B3PEI2 G3LF43 A0A2H1WEX3 A0A2A4J4V2 A0A2A4IW12 A0A0L7LM90 A0A3G1NI73 A0A194R7R7 I4DIH8 A0A194PW98 Q9U5K4 A0A1E1W5U0 A0A212ENI8 A0A1E1WLP6 A0A2H1WFB5 Q24995 A0A1E1WJ47 C6ETM8 Q9GQ56 Q9GQ55 A0A088G3X5 Q0EAG4 D3XLC1 A0A1Z2RRG4 Q0WYG7 Q0EAG5 A4GTP1 Q16XN3 A0A182GR26 B0WKQ2 A0A182F863 A0A182FQC3 W5J942 Q16I89 W5JSY3 Q16QU8 A0A1S4FSI4 A0A182H457 T1DNH0 B0W460 A0A182H055 A0A182Q072 A0A182MT47 Q7PUR0 A0A182X9P1 A0A182TGU7 A0A182L8X1 A0A182HKM5 A0A182R4M0 T1EA57 A0A182UUP7 A0A182Y3M6 P90664 A0A182PGJ5 A0A182NS01 A0A182VLH6 A0A182RZ96 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182WSL9 A0A182VZG2 Q7PXY7 A0A182JQ42 A0A182YGB4 A0A182VR52 A0A182MBJ5 A0A182J7X8 A0A182IRZ0 A0A182P2L1 A0A182KE62 A0A182S9W9 A0A084VS76 B0W462 A0A1B0D919 A0A1B0GLC3 A0A182Q5V2 A0A182HAP9 Q16I87 A0A1S4FZY0 A0A1J1J6D1 A0A182NSB7 A0A182IZN6 A0A084WBS9 A0A182QUT9
P14297 U3KV76 A0A2W1BLW4 U3KV63 Q7Z1F8 A0A3G1NIA0 A0A2W1BJI5 P20613 A0A1B3PEI2 G3LF43 A0A2H1WEX3 A0A2A4J4V2 A0A2A4IW12 A0A0L7LM90 A0A3G1NI73 A0A194R7R7 I4DIH8 A0A194PW98 Q9U5K4 A0A1E1W5U0 A0A212ENI8 A0A1E1WLP6 A0A2H1WFB5 Q24995 A0A1E1WJ47 C6ETM8 Q9GQ56 Q9GQ55 A0A088G3X5 Q0EAG4 D3XLC1 A0A1Z2RRG4 Q0WYG7 Q0EAG5 A4GTP1 Q16XN3 A0A182GR26 B0WKQ2 A0A182F863 A0A182FQC3 W5J942 Q16I89 W5JSY3 Q16QU8 A0A1S4FSI4 A0A182H457 T1DNH0 B0W460 A0A182H055 A0A182Q072 A0A182MT47 Q7PUR0 A0A182X9P1 A0A182TGU7 A0A182L8X1 A0A182HKM5 A0A182R4M0 T1EA57 A0A182UUP7 A0A182Y3M6 P90664 A0A182PGJ5 A0A182NS01 A0A182VLH6 A0A182RZ96 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182WSL9 A0A182VZG2 Q7PXY7 A0A182JQ42 A0A182YGB4 A0A182VR52 A0A182MBJ5 A0A182J7X8 A0A182IRZ0 A0A182P2L1 A0A182KE62 A0A182S9W9 A0A084VS76 B0W462 A0A1B0D919 A0A1B0GLC3 A0A182Q5V2 A0A182HAP9 Q16I87 A0A1S4FZY0 A0A1J1J6D1 A0A182NSB7 A0A182IZN6 A0A084WBS9 A0A182QUT9
PDB
3WJM
E-value=0,
Score=3213
Ontologies
PANTHER
Topology
Subcellular location
SignalP
Position: 1 - 16,
Likelihood: 0.996959
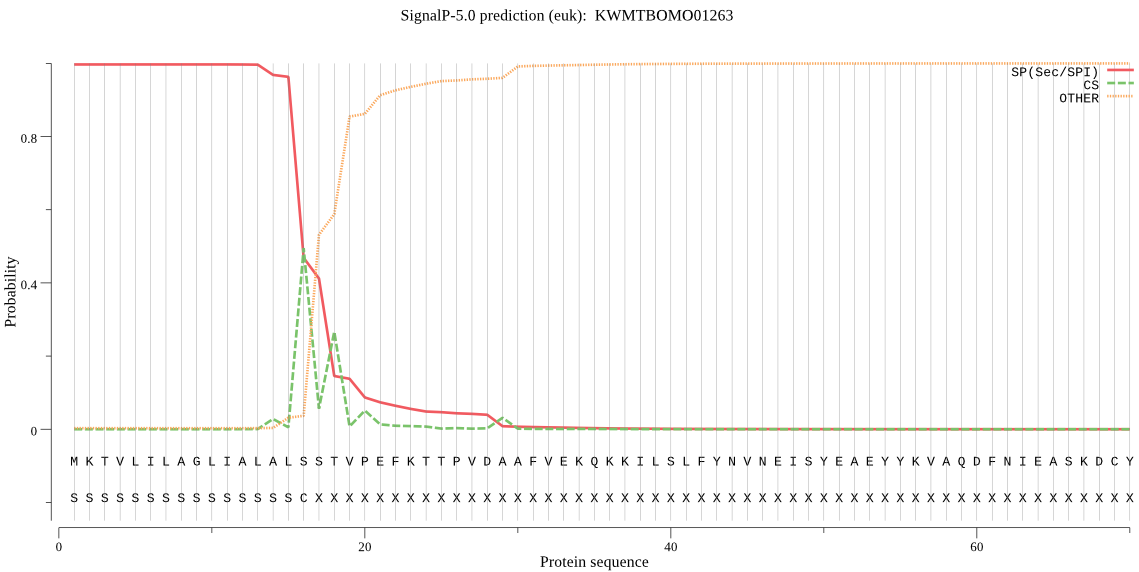
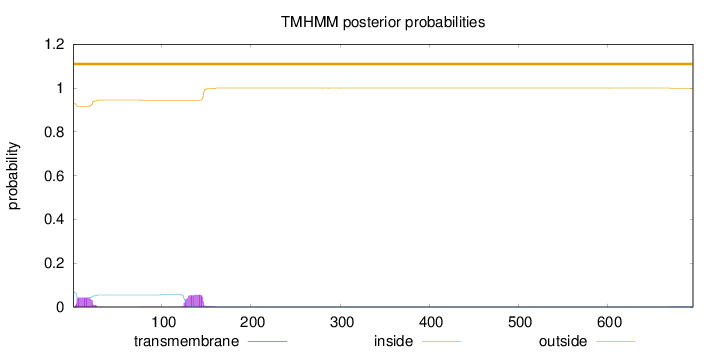
Length:
696
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.95271
Exp number, first 60 AAs:
0.77563
Total prob of N-in:
0.07034
outside
1 - 696
Population Genetic Test Statistics
Pi
171.983001
Theta
166.136522
Tajima's D
-1.36562
CLR
2.340953
CSRT
0.0786960651967402
Interpretation
Uncertain
