Gene
KWMTBOMO01256
Annotation
PREDICTED:_synaptotagmin-15-like_[Bombyx_mori]
Full name
Synaptotagmin-15
Alternative Name
Synaptotagmin XV
Chr10Syt
Chr10Syt
Location in the cell
Mitochondrial Reliability : 1.841 Nuclear Reliability : 1.769
Sequence
CDS
ATGATACTCGGTCGCGTGTGGTTCACGTTGAAGTACGAAGCAGCCACTGAAAGACTACAAGTGCATATCATAAAAGCTAAGCATCTGCCCTCACGTACACCGGCCCTGGCTAACGCATGCGATCCTTACATCAAGATTCAACTGATGCCCGACGAAAGGCGTGTTTTGCAAACGAAACAAAAGAAGAAAACCTGCAATCCTTTCTACGACGAAACTTTTGTTTACCAAATACCGCCGGGCAAAATTGACGCTCTCACCTTGAAGCTATCAGTAGTTGATGTGGGTAGGGTCGGGAAGGGAAAGCAGCTGATTGGACACATCATACTCCCATTGAGCGAATTGGACGGAGTGACACCCGACGAAGAACCACAATTCTACAAGCTTGATATCGAAAAGGACTTGCAAGAACCAACGTCAGATCTTGGTGAGATCCTGGTGTCTTTGCTATACAATGAAAATCTTCATCGACTCACAGTCACCGTCATCGAAGCGCGAGGGCTTAAACTGGATCCGTCGTCGAGCAAGCGTGAGATGGTAGTACGTGTAACTCAAGAGCGTGCTTGCCGCGGAGTCCGAGCAAGGAGGACGGCTGCTGTGGAAGTCACTGATGACGGTAGCTGCTTAGTCTCAGAGTGCTTCCACTTCCGGCTGCGAGCCGACGAACTTCACACCACGAGTGTTACCGTACAAGCCCTACAACCACACTCTGTCTATGCAAAAGACCGCCTTCTCGGTAAGTTTGTCCTAGGCTCGTACATGTTCGCTCGCGGCCGAGCCCTCCAACATTGGAACTCTGCGCTGGCATCGCCGATGGAACAAGTCAAACAATGGCACCCGCTCACCAACTAA
Protein
MILGRVWFTLKYEAATERLQVHIIKAKHLPSRTPALANACDPYIKIQLMPDERRVLQTKQKKKTCNPFYDETFVYQIPPGKIDALTLKLSVVDVGRVGKGKQLIGHIILPLSELDGVTPDEEPQFYKLDIEKDLQEPTSDLGEILVSLLYNENLHRLTVTVIEARGLKLDPSSSKREMVVRVTQERACRGVRARRTAAVEVTDDGSCLVSECFHFRLRADELHTTSVTVQALQPHSVYAKDRLLGKFVLGSYMFARGRALQHWNSALASPMEQVKQWHPLTN
Summary
Description
May be involved in the trafficking and exocytosis of secretory vesicles in non-neuronal tissues.
Subunit
Homodimer.
Similarity
Belongs to the synaptotagmin family.
Keywords
Complete proteome
Membrane
Reference proteome
Repeat
Signal-anchor
Transmembrane
Transmembrane helix
Alternative splicing
Cell membrane
Feature
chain Synaptotagmin-15
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A2W1B8S8
A0A2H1WTL0
A0A194R8Q6
A0A194Q0W0
A0A2A4IWV9
A0A2J7RQV7
+ More
A0A1Y1N1B5 A0A1B6CT46 A0A1Y1MZD4 A0A1B6KAZ3 A0A0K8SWB9 X1YW52 R7V3D6 A0A210PFA0 A0A2G8JHA7 A0A1V9XI85 A0A1S3INI9 A0A1S3IQJ0 A0A1B6MT16 T1KC97 K1PWY9 V3ZE58 W4ZIW6 A0A147BX78 E9H1H2 A0A162SKG5 A0A0P4YRB5 E0VX47 A0A2P4SPK6 A0A091V2V3 A0A0A0B1M6 A0A091SVH2 A0A093FTJ3 A0A099YT25 A0A3Q0H8J0 A0A218UJ09 A0A093QVJ1 A0A093H3Z2 A0A3Q0H552 A0A151MTB0 A0A091NNA5 A0A3L8SN89 A0A091T7Z4 A0A1D5NVT5 A0A093SSZ9 H0Z5X5 A0A091UI38 A0A091F763 M3VUI3 U3JW73 F1P2U3 A0A1L1RMJ9 F7CEC4 G1MU16 A0A3M0KX74 A0A087QH98 A0A2I0MFL8 A0A2I0MFK8 A0A094KIZ4 M7B002 A0A1D1V6Y5 A0A1V4KJH7 A0A1V4KJQ9 A0A1V4KJL3 U6D053 G9KS60 F6UWC2 A0A1L8FK77 A0A1W0WHQ7 G7N1Z5 G7PEV1 A0A2K6L1Z3 A0A2K6NUM7 G3W338 M3YC29 K7FDG6 A0A091PPA1 A0A226EYM2 A0A096P4C5 A0A2K5WBL4 Q5XFY6 A0A2K5IBJ3 A0A2K5M522 A0A0D9RBY9 F7EXZ9 D2H1K7 P59926 G3RX04 A0A2U3ZQJ7 Q9BQS2 A0A091H3Y4 A0A2I3RJ29 A0A093GNC8 A0A2K6BPJ7 A0A2Y9JDI6 F6RX20 V8P4U3 A0A096LNH4
A0A1Y1N1B5 A0A1B6CT46 A0A1Y1MZD4 A0A1B6KAZ3 A0A0K8SWB9 X1YW52 R7V3D6 A0A210PFA0 A0A2G8JHA7 A0A1V9XI85 A0A1S3INI9 A0A1S3IQJ0 A0A1B6MT16 T1KC97 K1PWY9 V3ZE58 W4ZIW6 A0A147BX78 E9H1H2 A0A162SKG5 A0A0P4YRB5 E0VX47 A0A2P4SPK6 A0A091V2V3 A0A0A0B1M6 A0A091SVH2 A0A093FTJ3 A0A099YT25 A0A3Q0H8J0 A0A218UJ09 A0A093QVJ1 A0A093H3Z2 A0A3Q0H552 A0A151MTB0 A0A091NNA5 A0A3L8SN89 A0A091T7Z4 A0A1D5NVT5 A0A093SSZ9 H0Z5X5 A0A091UI38 A0A091F763 M3VUI3 U3JW73 F1P2U3 A0A1L1RMJ9 F7CEC4 G1MU16 A0A3M0KX74 A0A087QH98 A0A2I0MFL8 A0A2I0MFK8 A0A094KIZ4 M7B002 A0A1D1V6Y5 A0A1V4KJH7 A0A1V4KJQ9 A0A1V4KJL3 U6D053 G9KS60 F6UWC2 A0A1L8FK77 A0A1W0WHQ7 G7N1Z5 G7PEV1 A0A2K6L1Z3 A0A2K6NUM7 G3W338 M3YC29 K7FDG6 A0A091PPA1 A0A226EYM2 A0A096P4C5 A0A2K5WBL4 Q5XFY6 A0A2K5IBJ3 A0A2K5M522 A0A0D9RBY9 F7EXZ9 D2H1K7 P59926 G3RX04 A0A2U3ZQJ7 Q9BQS2 A0A091H3Y4 A0A2I3RJ29 A0A093GNC8 A0A2K6BPJ7 A0A2Y9JDI6 F6RX20 V8P4U3 A0A096LNH4
Pubmed
28756777
26354079
28004739
23254933
28812685
29023486
+ More
28327890 22992520 29652888 21292972 20566863 22293439 30282656 20360741 17975172 15592404 17495919 20838655 23371554 23624526 27649274 23236062 20431018 27762356 22002653 25362486 21709235 17381049 15489334 15057822 15632090 17431167 12788067 14702039 15164054 11543631 16136131 24297900
28327890 22992520 29652888 21292972 20566863 22293439 30282656 20360741 17975172 15592404 17495919 20838655 23371554 23624526 27649274 23236062 20431018 27762356 22002653 25362486 21709235 17381049 15489334 15057822 15632090 17431167 12788067 14702039 15164054 11543631 16136131 24297900
EMBL
KZ150348
PZC71271.1
ODYU01010965
SOQ56400.1
KQ460597
KPJ13884.1
+ More
KQ459591 KPI97040.1 NWSH01005316 PCG64295.1 NEVH01000618 PNF43215.1 GEZM01020176 JAV89337.1 GEDC01020731 JAS16567.1 GEZM01020177 JAV89336.1 GEBQ01031361 JAT08616.1 GBRD01008234 JAG57587.1 AMQN01000901 KB297234 ELU10846.1 NEDP02076739 OWF35164.1 MRZV01001987 PIK35099.1 MNPL01010546 OQR73062.1 GEBQ01000895 JAT39082.1 CAEY01001958 JH818208 EKC20900.1 KB202619 ESO89388.1 AAGJ04141335 GEGO01000051 JAR95353.1 GL732583 EFX74350.1 LRGB01000024 KZS21390.1 GDIP01224926 JAI98475.1 DS235829 EEB17953.1 PPHD01030831 POI26036.1 KL410604 KFQ97651.1 KL873655 KGL99683.1 KK488411 KFQ62737.1 KK638924 KFV59995.1 KL885928 KGL73429.1 MUZQ01000269 OWK53714.1 KL224692 KFW62719.1 KL205760 KFV74135.1 AKHW03005127 KYO27742.1 KL387486 KFP90419.1 QUSF01000014 RLW04238.1 KK442201 KFQ70554.1 KL672327 KFW85639.1 ABQF01025298 KK434476 KFQ89857.1 KK719753 KFO65993.1 AANG04000273 AGTO01010261 AADN05000334 QRBI01000108 RMC11717.1 KL225592 KFM00602.1 AKCR02000016 PKK28474.1 PKK28473.1 KL260302 KFZ59273.1 KB560900 EMP28730.1 BDGG01000004 GAU97449.1 LSYS01003057 OPJ84604.1 OPJ84603.1 OPJ84605.1 HAAF01004089 CCP75914.1 JP019141 AES07739.1 AAMC01118624 AAMC01118625 CM004478 OCT71983.1 MTYJ01000100 OQV14712.1 CM001261 EHH19022.1 CM001284 EHH64695.1 AEFK01003550 AEFK01003551 AEFK01003552 AEYP01064600 AGCU01014853 AGCU01014854 KK676729 KFQ09480.1 LNIX01000001 OXA62702.1 AHZZ02035252 AQIA01070993 AABR07024658 BC084685 CH474046 AAH84685.1 EDL88884.1 AQIB01149122 JSUE03043359 GL192429 EFB16135.1 AB109021 CABD030071029 CABD030071030 AB109022 AB109023 AK127436 AK131036 AL356056 BC139914 AJ303363 KL448184 KFO81037.1 AACZ04003275 AACZ04043568 NBAG03000770 PNI11427.1 KL216210 KFV68294.1 AZIM01000864 ETE69023.1 FO681492
KQ459591 KPI97040.1 NWSH01005316 PCG64295.1 NEVH01000618 PNF43215.1 GEZM01020176 JAV89337.1 GEDC01020731 JAS16567.1 GEZM01020177 JAV89336.1 GEBQ01031361 JAT08616.1 GBRD01008234 JAG57587.1 AMQN01000901 KB297234 ELU10846.1 NEDP02076739 OWF35164.1 MRZV01001987 PIK35099.1 MNPL01010546 OQR73062.1 GEBQ01000895 JAT39082.1 CAEY01001958 JH818208 EKC20900.1 KB202619 ESO89388.1 AAGJ04141335 GEGO01000051 JAR95353.1 GL732583 EFX74350.1 LRGB01000024 KZS21390.1 GDIP01224926 JAI98475.1 DS235829 EEB17953.1 PPHD01030831 POI26036.1 KL410604 KFQ97651.1 KL873655 KGL99683.1 KK488411 KFQ62737.1 KK638924 KFV59995.1 KL885928 KGL73429.1 MUZQ01000269 OWK53714.1 KL224692 KFW62719.1 KL205760 KFV74135.1 AKHW03005127 KYO27742.1 KL387486 KFP90419.1 QUSF01000014 RLW04238.1 KK442201 KFQ70554.1 KL672327 KFW85639.1 ABQF01025298 KK434476 KFQ89857.1 KK719753 KFO65993.1 AANG04000273 AGTO01010261 AADN05000334 QRBI01000108 RMC11717.1 KL225592 KFM00602.1 AKCR02000016 PKK28474.1 PKK28473.1 KL260302 KFZ59273.1 KB560900 EMP28730.1 BDGG01000004 GAU97449.1 LSYS01003057 OPJ84604.1 OPJ84603.1 OPJ84605.1 HAAF01004089 CCP75914.1 JP019141 AES07739.1 AAMC01118624 AAMC01118625 CM004478 OCT71983.1 MTYJ01000100 OQV14712.1 CM001261 EHH19022.1 CM001284 EHH64695.1 AEFK01003550 AEFK01003551 AEFK01003552 AEYP01064600 AGCU01014853 AGCU01014854 KK676729 KFQ09480.1 LNIX01000001 OXA62702.1 AHZZ02035252 AQIA01070993 AABR07024658 BC084685 CH474046 AAH84685.1 EDL88884.1 AQIB01149122 JSUE03043359 GL192429 EFB16135.1 AB109021 CABD030071029 CABD030071030 AB109022 AB109023 AK127436 AK131036 AL356056 BC139914 AJ303363 KL448184 KFO81037.1 AACZ04003275 AACZ04043568 NBAG03000770 PNI11427.1 KL216210 KFV68294.1 AZIM01000864 ETE69023.1 FO681492
Proteomes
UP000053240
UP000053268
UP000218220
UP000235965
UP000014760
UP000242188
+ More
UP000230750 UP000192247 UP000085678 UP000015104 UP000005408 UP000030746 UP000007110 UP000000305 UP000076858 UP000009046 UP000053283 UP000053858 UP000053641 UP000189705 UP000197619 UP000054081 UP000053584 UP000050525 UP000276834 UP000053258 UP000007754 UP000052976 UP000011712 UP000016665 UP000000539 UP000002280 UP000001645 UP000269221 UP000053286 UP000053872 UP000031443 UP000186922 UP000190648 UP000008143 UP000186698 UP000009130 UP000233180 UP000233200 UP000007648 UP000000715 UP000007267 UP000198287 UP000028761 UP000233100 UP000002494 UP000233080 UP000233060 UP000029965 UP000006718 UP000001519 UP000245340 UP000005640 UP000053760 UP000002277 UP000053875 UP000233120 UP000248482 UP000008225
UP000230750 UP000192247 UP000085678 UP000015104 UP000005408 UP000030746 UP000007110 UP000000305 UP000076858 UP000009046 UP000053283 UP000053858 UP000053641 UP000189705 UP000197619 UP000054081 UP000053584 UP000050525 UP000276834 UP000053258 UP000007754 UP000052976 UP000011712 UP000016665 UP000000539 UP000002280 UP000001645 UP000269221 UP000053286 UP000053872 UP000031443 UP000186922 UP000190648 UP000008143 UP000186698 UP000009130 UP000233180 UP000233200 UP000007648 UP000000715 UP000007267 UP000198287 UP000028761 UP000233100 UP000002494 UP000233080 UP000233060 UP000029965 UP000006718 UP000001519 UP000245340 UP000005640 UP000053760 UP000002277 UP000053875 UP000233120 UP000248482 UP000008225
Pfam
PF00168 C2
Gene 3D
ProteinModelPortal
A0A2W1B8S8
A0A2H1WTL0
A0A194R8Q6
A0A194Q0W0
A0A2A4IWV9
A0A2J7RQV7
+ More
A0A1Y1N1B5 A0A1B6CT46 A0A1Y1MZD4 A0A1B6KAZ3 A0A0K8SWB9 X1YW52 R7V3D6 A0A210PFA0 A0A2G8JHA7 A0A1V9XI85 A0A1S3INI9 A0A1S3IQJ0 A0A1B6MT16 T1KC97 K1PWY9 V3ZE58 W4ZIW6 A0A147BX78 E9H1H2 A0A162SKG5 A0A0P4YRB5 E0VX47 A0A2P4SPK6 A0A091V2V3 A0A0A0B1M6 A0A091SVH2 A0A093FTJ3 A0A099YT25 A0A3Q0H8J0 A0A218UJ09 A0A093QVJ1 A0A093H3Z2 A0A3Q0H552 A0A151MTB0 A0A091NNA5 A0A3L8SN89 A0A091T7Z4 A0A1D5NVT5 A0A093SSZ9 H0Z5X5 A0A091UI38 A0A091F763 M3VUI3 U3JW73 F1P2U3 A0A1L1RMJ9 F7CEC4 G1MU16 A0A3M0KX74 A0A087QH98 A0A2I0MFL8 A0A2I0MFK8 A0A094KIZ4 M7B002 A0A1D1V6Y5 A0A1V4KJH7 A0A1V4KJQ9 A0A1V4KJL3 U6D053 G9KS60 F6UWC2 A0A1L8FK77 A0A1W0WHQ7 G7N1Z5 G7PEV1 A0A2K6L1Z3 A0A2K6NUM7 G3W338 M3YC29 K7FDG6 A0A091PPA1 A0A226EYM2 A0A096P4C5 A0A2K5WBL4 Q5XFY6 A0A2K5IBJ3 A0A2K5M522 A0A0D9RBY9 F7EXZ9 D2H1K7 P59926 G3RX04 A0A2U3ZQJ7 Q9BQS2 A0A091H3Y4 A0A2I3RJ29 A0A093GNC8 A0A2K6BPJ7 A0A2Y9JDI6 F6RX20 V8P4U3 A0A096LNH4
A0A1Y1N1B5 A0A1B6CT46 A0A1Y1MZD4 A0A1B6KAZ3 A0A0K8SWB9 X1YW52 R7V3D6 A0A210PFA0 A0A2G8JHA7 A0A1V9XI85 A0A1S3INI9 A0A1S3IQJ0 A0A1B6MT16 T1KC97 K1PWY9 V3ZE58 W4ZIW6 A0A147BX78 E9H1H2 A0A162SKG5 A0A0P4YRB5 E0VX47 A0A2P4SPK6 A0A091V2V3 A0A0A0B1M6 A0A091SVH2 A0A093FTJ3 A0A099YT25 A0A3Q0H8J0 A0A218UJ09 A0A093QVJ1 A0A093H3Z2 A0A3Q0H552 A0A151MTB0 A0A091NNA5 A0A3L8SN89 A0A091T7Z4 A0A1D5NVT5 A0A093SSZ9 H0Z5X5 A0A091UI38 A0A091F763 M3VUI3 U3JW73 F1P2U3 A0A1L1RMJ9 F7CEC4 G1MU16 A0A3M0KX74 A0A087QH98 A0A2I0MFL8 A0A2I0MFK8 A0A094KIZ4 M7B002 A0A1D1V6Y5 A0A1V4KJH7 A0A1V4KJQ9 A0A1V4KJL3 U6D053 G9KS60 F6UWC2 A0A1L8FK77 A0A1W0WHQ7 G7N1Z5 G7PEV1 A0A2K6L1Z3 A0A2K6NUM7 G3W338 M3YC29 K7FDG6 A0A091PPA1 A0A226EYM2 A0A096P4C5 A0A2K5WBL4 Q5XFY6 A0A2K5IBJ3 A0A2K5M522 A0A0D9RBY9 F7EXZ9 D2H1K7 P59926 G3RX04 A0A2U3ZQJ7 Q9BQS2 A0A091H3Y4 A0A2I3RJ29 A0A093GNC8 A0A2K6BPJ7 A0A2Y9JDI6 F6RX20 V8P4U3 A0A096LNH4
PDB
6MTI
E-value=2.27308e-25,
Score=285
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Cell membrane
Cell membrane
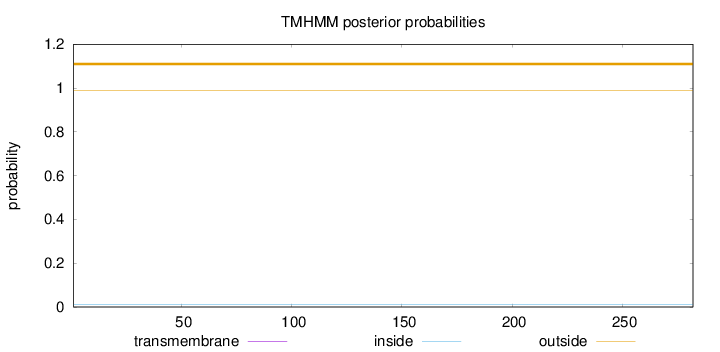
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00153
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01020
outside
1 - 282
Population Genetic Test Statistics
Pi
21.358322
Theta
183.30077
Tajima's D
0.752879
CLR
0.541707
CSRT
0.588770561471926
Interpretation
Uncertain
