Gene
KWMTBOMO01253
Pre Gene Modal
BGIBMGA007548
Annotation
PREDICTED:_uncharacterized_protein_LOC107191251_[Dufourea_novaeangliae]
Location in the cell
Nuclear Reliability : 3.879
Sequence
CDS
ATGATGAAGCACATGTCCGATCGTATGGCAGCAATACCGGACGCAAAAGTTAAAATCAATGACGTGTTTTTACCATCATACGATCCGGACGCAAATATAGGAGTGCGAGAGTGGTGTAAGCATGTGACCACGGCCATGGAAACCTACAACCTGAGCGATTATGATGTCAGAATGAAAGTCGGTAGTTTGCTCAAAGGTCGTGCAAGGTTGTGGGTTGACAACTGGCTTATCTCAACCTCCACGTGGCAGGAATTGCGTGATGTTATGATAACAACCTTTGAACCTGAAAATAGGTATTCTCGTGATATAGTTCGTTTTCGGGAACATAGTTACGATAATTCAAAGGACATCACGCAGTTCTTATCACAGGCTTGGATGTTATGGAGACGTGTCACTAAGGATAAATTATCGGATGATGACGCGGTTGAGGCTGTGATTGGTTGCATTGGAGATGAGCGATTGAGAATCGAGTTGTTAAACGCCAGGGCTACTTCGGTACCAGAGTTGATTTCTGTAGCTTCCTCAATAAAGCGCAGTAAGCGACCTTATCCTGGTTCATCAAATATGCAACAGGGCCCTGTCAAACGACAACGATTCTCGGATAAGCCGTCTTTATATTGTCAACAGTGTAAAAAGCCCGGTCATGATACTCGAGATTGCCGTTACCGTGATAAAGCTGATAACCCTCAACTGCGACATCATGATGATAAAACCGTTCCTCAAAGGGATAATAAACCGACCTGTACGTTCTGTTCACGAACAGGTCACACATATGAAACTTGTTATAAACGTGAACGAGCAATTGTCTCGAATGTTAATTGCGTAGGTGCACCTAAATTAAACTCCATACCGGTGTTGATTGGAGGTTTAAAATTTTTCGGGATTTTTGATAGCGGGGCGGAATGTTCGGTAATACGCGAATCGGTTGCTTCAAAATTACCGGGTAAAAGGACGAATGTAGTTAATTATCTAAAGGGCTTAGGACAGTTTACAGTAGTGTCATTATCAACGTTGATAACCGTTTGTGTGATCGATGATATAAGGGTCGAACTTCATTTCCACATAGTCCCGGATTATGAAGTGTGTTCAGATATTTTAATAGGCATGAATCTTATGGAAAATACAAATTTAAGTGTGATAGTGAATTCAAAGGGCGTGGCATTGATTCATCAGCCCGCGATTTTCCACATGCGATTACAAAGCGAAAAGTTTGACAATTTGGATTGTGACTTGACGGATAGTGATCAAATAAACGAACTTAAGTTGCTATTAGCAAAATTTGAACATATTTTTATCCAGGGTTACCCTCACACGCGTGTTAATACCGGTGAACTCGAAATACGTCTGAAAGACCCCAACAAGTGTGTTGAGCGTAGGCCATATAGATTGAGTCCAATCGAACGACAAAAAGTTCGTGATATTGTGAGCGAGTTACTTGAACACAATATTATTCGAGAGAGCAAGTCACCTTTCTCCAGTCCGATAATATTAGTCAAAAAGAAAAACGGCAAAGATCGCATGTGCGTGGACTATCGCGAGCTGAATAGAAACACTCTTAGGGATCACTATCCTCTGCCTATAATTTCCGATCAAATAGACCAATTAGCGGGTGGTTATTACTTCTCTTCTTTTGATATGGCAGCAGGCTTTCATCAGATCCCCATTTCTAAAGGGTCTATTGAAAAGACTGCCTTCGTAACACCGGACGGCATGTACGAGTACTTGACCATGCCATTCGGTCTCAGTAATGCATGTTCCGTTTACCAGAGGTGCATGAATAGGGCTTTAGCTGACCTTCTAAATTCACCTGACCAGGTCTGTCAAGTGTACGTCGACGACGTACTAACAAAATGTTGTGAATTTACAGAAGGTTTGTCCCGTATCGAACGGGTACTTATCGCATTACAAGATGCCGGGTTTTCCATAAATATTGAGAAAACTGCCTTCTTTAAACGCTCTATCGAGTACTTGGGCAATATAGTTACAAACGGACAGGTTAGTCCTAGCCCAAAGAAAGTGGAGGCTTTAACGAAGGCACCTATTCCACGGACGGCTAAACAAGTAAGGCAATTCAATGGCCTCGCCGGATATTTTCGTCGTTTCATTCCCAATTTCTCGCGTGTGATGGTACCGCTTTATGAGCTAACAAAAAAAGACGCAAAATGGGAATGGAATGAAAGGCATGATGAGGCCAGAAATATAATCATACTGCATTTGTCTACTGCGCCTACCCTTGCATTATTTCAGGAAAACGCACCAGTTGAGCTTTACACAGATGCGAGCAGCCTCGGTTACGGCGCGGTACTGATTCAGATAATTGGAGGACGCCAGCATCCAGTCGCGTACATGAGCCAGCGTACCACTGACGCAGAGAGCCGCTACCATTCATATGAACTCGAGACGCTAGCAGTGGTGAGAGCAGTAAAACATTTTCGGCACTACCTATACGGCCGAAAATTCAAGATAATAACAGACTGTAACGCCTTGAAAGCATCTAAGCATAAGAAGGATTTACTGCCACGAATTCATCGCTGGTGGGCTTTTCTGCAGAACTACGAGTTCGAGGTCGAATATAGGAAAGGTGAGCGACTGCAGCACGCAGACTTTTTTAGCCGAAACCCTACAACTAAAATGGCGATCAACATCATGACCAAGGACGCAGAGTGGCTGCAAATTGAACAGCGTCGTGACGATACTCTTCGTCCTGTGATAAACAGCATGACAACCGACAACCCAACTCCCGGCTACGTTCTTGAGGAAGGCGTGCTGAAGAAATTACTCGCTGATCCTTCCATACTCGGTGCAATCCAGGCACAGCTCCATCCAATACAAAAACCTACTGCAGCGTTTCAAGTAGTCCACATAGACGTTACCGGAAAACTTGGTACGAGAAACTCTGAAGGTCAGGACGAATATGTGATAGTTATCATTGACGCCTTTACCAAATACATCCTACTTAGTTACTCTAACGACAAGAGTCAAAGCAGCAGTCTCGCAGCGCTGAAACGAGTCGTGCATTTGTTCGGGACCCCAGTCCAGGTAGTGGTTGATGGCGGCCGAGAATTTCTCGGCGAATTCAAAGCCTATTGCGACTGTTTTGGTATCAATTTACATGCTATAGCGCCAGGAGTAAGCCGAGCAAACGGGCAAGTGGAACGAATAATGAGTACCTTGAAAAATGCACTAACAATAATAAGAAATTATGACACTGAAAACTGGCAAACGGCCTTGGAAGCCCTTCAACTTGCATTTAATTGCACCCCACACAGAGTGACAGGGTGCGCGCCACTTACTCTCCTGACACGCCGTAAGCACACCGTTCCGCCAGAACTTTTAAGGTTGGTAGACATTGATAGCGAAACCGTCGATTTTGATATGCTCGATCAACACGTGCAACAGAAAATGGCGGCTGCAGCTGAATATGATAAGCATCGTTTTAACAGAAACAAAGCCAAGCTGCGTCCTTTTCAAAGAGGAGACTACGTCCTCATCAAAAACAACCCCAGAAATCAAACCAGCTTGGACCTGAAATACAGCGAGCCATACGAGATTTACAGAATAATGGACAATGACCGCTACATGGTAAAACGTGTGACCGGCAGAGGGCGCCCACGCAAGGTGGCGCACGATCAGCTGCGCCGGGCTCCACAGCCTGGGGAACAGGAAACCGTGTCGACGGGCGCCAACGATGACACCGAACAGCAGACCACAGCCCAAGTAGTCGATACACCGGATCCACCATCAACGTTATTAGACGCTTAA
Protein
MMKHMSDRMAAIPDAKVKINDVFLPSYDPDANIGVREWCKHVTTAMETYNLSDYDVRMKVGSLLKGRARLWVDNWLISTSTWQELRDVMITTFEPENRYSRDIVRFREHSYDNSKDITQFLSQAWMLWRRVTKDKLSDDDAVEAVIGCIGDERLRIELLNARATSVPELISVASSIKRSKRPYPGSSNMQQGPVKRQRFSDKPSLYCQQCKKPGHDTRDCRYRDKADNPQLRHHDDKTVPQRDNKPTCTFCSRTGHTYETCYKRERAIVSNVNCVGAPKLNSIPVLIGGLKFFGIFDSGAECSVIRESVASKLPGKRTNVVNYLKGLGQFTVVSLSTLITVCVIDDIRVELHFHIVPDYEVCSDILIGMNLMENTNLSVIVNSKGVALIHQPAIFHMRLQSEKFDNLDCDLTDSDQINELKLLLAKFEHIFIQGYPHTRVNTGELEIRLKDPNKCVERRPYRLSPIERQKVRDIVSELLEHNIIRESKSPFSSPIILVKKKNGKDRMCVDYRELNRNTLRDHYPLPIISDQIDQLAGGYYFSSFDMAAGFHQIPISKGSIEKTAFVTPDGMYEYLTMPFGLSNACSVYQRCMNRALADLLNSPDQVCQVYVDDVLTKCCEFTEGLSRIERVLIALQDAGFSINIEKTAFFKRSIEYLGNIVTNGQVSPSPKKVEALTKAPIPRTAKQVRQFNGLAGYFRRFIPNFSRVMVPLYELTKKDAKWEWNERHDEARNIIILHLSTAPTLALFQENAPVELYTDASSLGYGAVLIQIIGGRQHPVAYMSQRTTDAESRYHSYELETLAVVRAVKHFRHYLYGRKFKIITDCNALKASKHKKDLLPRIHRWWAFLQNYEFEVEYRKGERLQHADFFSRNPTTKMAINIMTKDAEWLQIEQRRDDTLRPVINSMTTDNPTPGYVLEEGVLKKLLADPSILGAIQAQLHPIQKPTAAFQVVHIDVTGKLGTRNSEGQDEYVIVIIDAFTKYILLSYSNDKSQSSSLAALKRVVHLFGTPVQVVVDGGREFLGEFKAYCDCFGINLHAIAPGVSRANGQVERIMSTLKNALTIIRNYDTENWQTALEALQLAFNCTPHRVTGCAPLTLLTRRKHTVPPELLRLVDIDSETVDFDMLDQHVQQKMAAAAEYDKHRFNRNKAKLRPFQRGDYVLIKNNPRNQTSLDLKYSEPYEIYRIMDNDRYMVKRVTGRGRPRKVAHDQLRRAPQPGEQETVSTGANDDTEQQTTAQVVDTPDPPSTLLDA
Summary
Uniprot
B7S8P8
A0A0A9WUU5
A0A0A9WTW7
Q24310
A0A0A9WJJ8
A0A0A9X1Z8
+ More
A0A146M2J2 A0A023EZG2 A0A023EYH9 A0A023EY26 J9JZ77 A0A1W7R6G2 A0A1W7R6L8 A0A0J7KLK4 A0A0J7NF11 X1X5H2 A0A2S2NKJ2 J9JQ65 A0A034WRX5 X1XU22 W8C8Q1 W8B0V9 W8APB2 J9KLX4 A0A034VAM6 A0A2M4AEU2 Q24262 A0A0J7K890 T1PMR9 A0A0A1WKC8 Q961S4 A0A034VPR8 A0A0J7K9X8 L7ML53 L7LV95 A0A023F013 A0A023EYF0
A0A146M2J2 A0A023EZG2 A0A023EYH9 A0A023EY26 J9JZ77 A0A1W7R6G2 A0A1W7R6L8 A0A0J7KLK4 A0A0J7NF11 X1X5H2 A0A2S2NKJ2 J9JQ65 A0A034WRX5 X1XU22 W8C8Q1 W8B0V9 W8APB2 J9KLX4 A0A034VAM6 A0A2M4AEU2 Q24262 A0A0J7K890 T1PMR9 A0A0A1WKC8 Q961S4 A0A034VPR8 A0A0J7K9X8 L7ML53 L7LV95 A0A023F013 A0A023EYF0
EMBL
EF710649
ACE75273.1
GBHO01032413
JAG11191.1
GBHO01032415
JAG11189.1
+ More
X14037 CAA32198.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GBHO01030786 JAG12818.1 GDHC01004980 JAQ13649.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 GBBI01004477 JAC14235.1 ABLF02014660 ABLF02042774 GEHC01000888 JAV46757.1 GEHC01000930 JAV46715.1 LBMM01005919 KMQ91096.1 KMQ91095.1 ABLF02004401 ABLF02013544 ABLF02013550 GGMR01005091 MBY17710.1 ABLF02005846 GAKP01001563 JAC57389.1 ABLF02017401 GAMC01007326 JAB99229.1 GAMC01015907 JAB90648.1 GAMC01020207 JAB86348.1 ABLF02017561 ABLF02041657 ABLF02042885 GAKP01019383 JAC39569.1 GGFK01005990 MBW39311.1 Z27119 CAA81643.1 LBMM01011782 KMQ86633.1 KA649989 AFP64618.1 GBXI01015151 JAC99140.1 AY050234 AAK84933.1 GAKP01014483 JAC44469.1 LBMM01010976 KMQ87102.1 GACK01000334 JAA64700.1 GACK01009043 JAA55991.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1
X14037 CAA32198.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GBHO01030786 JAG12818.1 GDHC01004980 JAQ13649.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 GBBI01004477 JAC14235.1 ABLF02014660 ABLF02042774 GEHC01000888 JAV46757.1 GEHC01000930 JAV46715.1 LBMM01005919 KMQ91096.1 KMQ91095.1 ABLF02004401 ABLF02013544 ABLF02013550 GGMR01005091 MBY17710.1 ABLF02005846 GAKP01001563 JAC57389.1 ABLF02017401 GAMC01007326 JAB99229.1 GAMC01015907 JAB90648.1 GAMC01020207 JAB86348.1 ABLF02017561 ABLF02041657 ABLF02042885 GAKP01019383 JAC39569.1 GGFK01005990 MBW39311.1 Z27119 CAA81643.1 LBMM01011782 KMQ86633.1 KA649989 AFP64618.1 GBXI01015151 JAC99140.1 AY050234 AAK84933.1 GAKP01014483 JAC44469.1 LBMM01010976 KMQ87102.1 GACK01000334 JAA64700.1 GACK01009043 JAA55991.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1
Proteomes
Pfam
Interpro
IPR041577
RT_RNaseH_2
+ More
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR019103 Peptidase_aspartic_DDI1-type
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR034145 RP_RTVL-H-like
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR019103 Peptidase_aspartic_DDI1-type
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR034145 RP_RTVL-H-like
Gene 3D
ProteinModelPortal
B7S8P8
A0A0A9WUU5
A0A0A9WTW7
Q24310
A0A0A9WJJ8
A0A0A9X1Z8
+ More
A0A146M2J2 A0A023EZG2 A0A023EYH9 A0A023EY26 J9JZ77 A0A1W7R6G2 A0A1W7R6L8 A0A0J7KLK4 A0A0J7NF11 X1X5H2 A0A2S2NKJ2 J9JQ65 A0A034WRX5 X1XU22 W8C8Q1 W8B0V9 W8APB2 J9KLX4 A0A034VAM6 A0A2M4AEU2 Q24262 A0A0J7K890 T1PMR9 A0A0A1WKC8 Q961S4 A0A034VPR8 A0A0J7K9X8 L7ML53 L7LV95 A0A023F013 A0A023EYF0
A0A146M2J2 A0A023EZG2 A0A023EYH9 A0A023EY26 J9JZ77 A0A1W7R6G2 A0A1W7R6L8 A0A0J7KLK4 A0A0J7NF11 X1X5H2 A0A2S2NKJ2 J9JQ65 A0A034WRX5 X1XU22 W8C8Q1 W8B0V9 W8APB2 J9KLX4 A0A034VAM6 A0A2M4AEU2 Q24262 A0A0J7K890 T1PMR9 A0A0A1WKC8 Q961S4 A0A034VPR8 A0A0J7K9X8 L7ML53 L7LV95 A0A023F013 A0A023EYF0
PDB
4OL8
E-value=9.95193e-70,
Score=674
Ontologies
GO
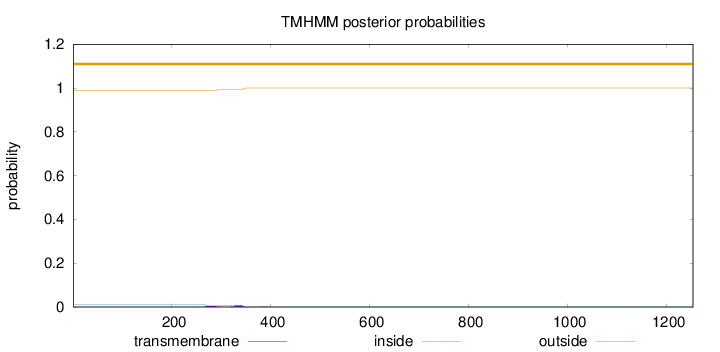
Topology
Length:
1253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26688
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01189
outside
1 - 1253
Population Genetic Test Statistics
Pi
33.917726
Theta
215.763695
Tajima's D
-1.72349
CLR
1362.583872
CSRT
0.0356482175891205
Interpretation
Uncertain
