Gene
KWMTBOMO01252
Pre Gene Modal
BGIBMGA008856
Annotation
PREDICTED:_cGMP-dependent_protein_kinase?_isozyme_2_forms_cD5/T2-like_isoform_X1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 4.363
Sequence
CDS
ATGAAAATCAAACATTATCCAGGAAAAGCGGTCATGGAGGACCCGTGGCTATCCCTCGGGGATAGAACTATGGACCTTGTCTATGCGGCAAACTGGGTCGGCGCTAAGAATCTTAATAAATGCAATTCTAACGACGATCAATCAAGTTCAAAATGTAATTCTATATCGAGGAGCAAATCATGTAGAGATGTCGGCCGTTCGGAAGAAAACGAATACAGCCAAAAAAAATACTCACAAGAAGACAGTGGGTGTTACTTTGATGTTCCTAATGAAAACATATCTCAATTAGCCAAATATCAACAAAATATAGAAGAAAAAGCTAGGACGAGTGATAATTACATTGCTCAAAACAAATTAAAGCACAGTTATAGTTTTAAAGATATGCCAAATGGCTATAAGCCTTCGACTCTGCGACGCGTGAAACGAAGAAATTTCACAGAATGGGACATCGAAGCATTTGGTTCGAATAGTAAACAGCCACCCGTTCCGCCACAACGTAAAGATTCTTTGAAATACGCACATCGACAGAGAAAAGTTGATTCAAAGAAAAATTACTATCCGTTACCTGATCCTGGTCCAGTACCTCCTGATCCATCATCAGAATCATATTATGAATATTATTCTAGAGTCAGTCAACAAAACAATGACACCGATAGCGATAAGCTAAAGCAGATCCAAAAAAGTGGTAAGAATGATAGAAATTCGCCAAATGAATACCCCAAACAAAACGGTAATACCGTGTACAAAATTATACATTCAATGGCCACCTTAGATCTGAACCCGATGAAAGTCAGAAACGGTAAACAAACTTTGGAATTGGATAGTGAACCGGAAGATTTGGGTCTATACATGCGGCCTATTAAACAGTCGTTATCTACATGCGATCAACTTCCGAACCACACTACGCAACCGGCGAACGAACGAAACGGACGCATCGAAGACGCTAAACCTAGCTTTCGTTCTAAATCATTAAGAGTTAAAAGCGAAGGTAGAGTACCACTTCGAGCGTACCTTGACACGGTCCAGCACAACAATGCGCAGTACAACGGTGTCACTGTCACGCAACCGAAAAGAAATCTGTCGCCGAGCGAACAGTTGACGCTAATGCAGTACGAAATGTTCAACGGAAGTAAGAACCAACTCGCACCGCCGATCAGCAACGGGTTCCACGCTGTTGATCCGTCTAAAGAAAGTGATTATCGGACTATAAATAGTGTTACAAAACAAATCAAACCCGATAAATATGGTAAGTTATATAACACCGTGAGAGTGAAAAGTGACGATAGATACGAAAGGTACAATGATATTACAAAATTCGCACCTCCGAAAGACCATATCAATGAGGAACGTGCAACTAATGGAAATGGTGTGTTTACGAGAAGTGCTAAAAGTTCATCTAGTGGTTCGGACTCTGATTTTTCAATACCACGGCCGAAATTAATAGTACCTGTTCATACCTATGGAACACGCAAGAGACGAACTGGGAATGTACAGCGTGATAGCACCGCAACGGAAGCTTCATTGGATGCGGGGATCGTGTTAGACGTGACACGATTGGAAGATTCGAGTCATAAGTTTAAGAAAAATGAAGAATCAGAAGGCCGAGTCGAAGTGTCGAGGGAGAACAAATACCTGAGCACGATGGCGCCAGGCAAGGTCTTCGGGGAACTGGCAATCCTGTACAACTGCAAGCGTACAGCTACCATCAAAGCCGCTACGGACTGCAGACTTTGGGCCATCGAACGACAATGCTTCCAAACGATTATGATGAGAACTGGCCTTATCAGACAGGCTGAATACACAGACTTCTTGAAGAGTGTACCGATCTTCAAGAACTTCCCCGAAGATACGTTGATTAAAATCTCCGATGTGTTGGAAGAGACTCATTACCAGAATGGAGACTATATTATTAGACAGGGTGCGAGAGGTGACACGTTCTTCATCATATCCAAAGGACAGGTTAAAGTTACACAAAAACTACCGAATAGTAACGACGAGAAATTCATCAGAACCCTCACGAAAGGAGACTTCTTCGGAGAGAAGGCCTTACAAGGGGACGACCTCCGAACGGCGAATATCGTCTGTGACTCACCAGACGGCACGACTTGTCTCGTTATAGACAGGGAAACATTCAATCAGCTGATATCCACACTGGACGAGATCCGAACCAAGTACAAGGATCTGGGTGATGACAGACAACGATTGAACGAAGAATTCGCCAACCTGCGTCTCTCGGATCTCCGCATTATCGCTACATTGGGTATCGGAGGCTTCGGTCGAGTGGAATTAGTGCAAATCCAAGGCGATTCGAGTCGCTCCTTCGCCTTAAAGCAGATGAAGAAGGCCCAGATCGTTGAGACGAGACAACAACAACACATAATGTCCGAAAAGGAGATAATGTCCGAAATGAACTGCGAATTCATAGTGAAGCTATACAAGACTTTCAAAGATCGCAAGTACTTGTACATGTTGATGGAGACTTGCCTCGGTGGAGAGCTATGGACCATTTTAAGAGATCGAGGACAATTTGACGACGCAACGACAAGATTCTACACGGCCTGTGTAGTTGAGGCCTTCCACTATCTACATTCTAGAAATATAATTTACAGGGATCTCAAACCAGAGAATCTACTGCTAGACTCGAAAGGTTACGTGAAACTGGTCGACTTCGGTTTCTCAAAGAAGCTGCAGGCGAGCCGTAAGACCTGGACGTTCTGCGGCACGCCAGAGTACGTCGCGCCCGAGGTCATCATGAACCGGGGCCACGATATCAGTGCGGACTATTGGTCTTTAGGTGTTCTCATGTTCGAACTGCTGACCGGGTCGCCGCCCTTCACGGGCTCGGATCCTATGAAGACGTACAACAAAATCCTGAAGGGCATCGATGCGGTCGAGTTCCCGCGCTGCATCACGCGCAACGCCGCTAATCTGATCAAGAAACTCTGCCGCGACAACCCTGCGGAGCGGCTCGGCTATCAACGAGGCGGCATAACTGAGATACAGAAGCACAAATGGTTCGATGGCTTTAACTGGGAAGGCCTAGCGCAACGTTCTCTAGAGCCACCTATTGTACCGACCGTAAATTCGGCCATAGACACGCACAACTTCGACCAATACCCGGCAGATGCAGACGAACCTCCCCCCGACGATCTCTCCGGTTGGGATGCGAACTTTTAA
Protein
MKIKHYPGKAVMEDPWLSLGDRTMDLVYAANWVGAKNLNKCNSNDDQSSSKCNSISRSKSCRDVGRSEENEYSQKKYSQEDSGCYFDVPNENISQLAKYQQNIEEKARTSDNYIAQNKLKHSYSFKDMPNGYKPSTLRRVKRRNFTEWDIEAFGSNSKQPPVPPQRKDSLKYAHRQRKVDSKKNYYPLPDPGPVPPDPSSESYYEYYSRVSQQNNDTDSDKLKQIQKSGKNDRNSPNEYPKQNGNTVYKIIHSMATLDLNPMKVRNGKQTLELDSEPEDLGLYMRPIKQSLSTCDQLPNHTTQPANERNGRIEDAKPSFRSKSLRVKSEGRVPLRAYLDTVQHNNAQYNGVTVTQPKRNLSPSEQLTLMQYEMFNGSKNQLAPPISNGFHAVDPSKESDYRTINSVTKQIKPDKYGKLYNTVRVKSDDRYERYNDITKFAPPKDHINEERATNGNGVFTRSAKSSSSGSDSDFSIPRPKLIVPVHTYGTRKRRTGNVQRDSTATEASLDAGIVLDVTRLEDSSHKFKKNEESEGRVEVSRENKYLSTMAPGKVFGELAILYNCKRTATIKAATDCRLWAIERQCFQTIMMRTGLIRQAEYTDFLKSVPIFKNFPEDTLIKISDVLEETHYQNGDYIIRQGARGDTFFIISKGQVKVTQKLPNSNDEKFIRTLTKGDFFGEKALQGDDLRTANIVCDSPDGTTCLVIDRETFNQLISTLDEIRTKYKDLGDDRQRLNEEFANLRLSDLRIIATLGIGGFGRVELVQIQGDSSRSFALKQMKKAQIVETRQQQHIMSEKEIMSEMNCEFIVKLYKTFKDRKYLYMLMETCLGGELWTILRDRGQFDDATTRFYTACVVEAFHYLHSRNIIYRDLKPENLLLDSKGYVKLVDFGFSKKLQASRKTWTFCGTPEYVAPEVIMNRGHDISADYWSLGVLMFELLTGSPPFTGSDPMKTYNKILKGIDAVEFPRCITRNAANLIKKLCRDNPAERLGYQRGGITEIQKHKWFDGFNWEGLAQRSLEPPIVPTVNSAIDTHNFDQYPADADEPPPDDLSGWDANF
Summary
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
6C0T
E-value=2.75497e-138,
Score=1265
Ontologies
GO
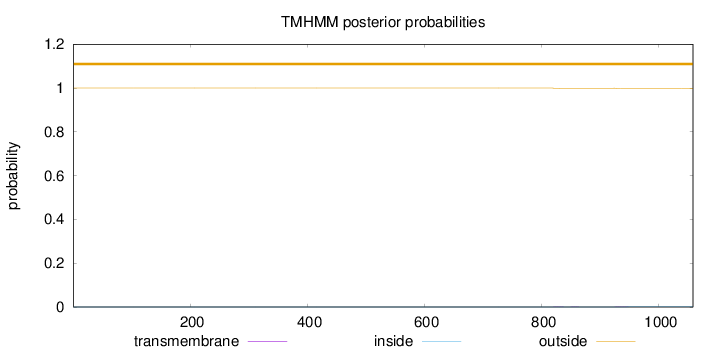
Topology
Length:
1058
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03042
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1058
Population Genetic Test Statistics
Pi
220.025937
Theta
187.970815
Tajima's D
0.687367
CLR
0.194112
CSRT
0.575671216439178
Interpretation
Uncertain
