Gene
KWMTBOMO01251
Pre Gene Modal
BGIBMGA008855
Annotation
cGMP-dependent_protein_kinase?_isozyme_2_forms_cD4/T1/T3A/T3B_[Papilio_xuthus]
Full name
cGMP-dependent protein kinase
+ More
Mediator of RNA polymerase II transcription subunit 4
Mediator of RNA polymerase II transcription subunit 4
Alternative Name
Mediator complex subunit 4
Location in the cell
Nuclear Reliability : 3.507
Sequence
CDS
ATGTCGCACACGCACACGAAACAGTCCCAAATGGAGCACAATTACAAATTGGAGAAGAAATATTTGGAAATGAAGCGAATGGTTTACGAACGTACCGAAGAACTAAAACGGAGGGATAAAATTATTACACTATTGGAAAACGAGATTGACGAGAAAGACGCTCAAATAAGATATCTCAAGAACGAGATTGACAAATATAGGCAAGTCGTGAAACCATTGACGCGTCAGATTATTGACAGCCACAAAAACGGAATCGATACCGACGAGGAATGCTTCACGATGAAGACTCCGGGCGTCGAGAATACGCGAGTTTATTCATTAAATGAACCACGACTGAAAAGGACTGCGATATCAGCGGAGCCCCTATCGTCGCTGACTCAAGAGCATGAGTTGAAGTTATTGAAAATACCGAAAAGCCACAGATCTCGCGAGCTGATTAAGACTGCGATCCTCGACAACGACTTCATGAAGAATCTAGAAATGACACAGATCAGAGAAATAGTGGATTGCATGTATCCCGTCGAATATGCAGCCGGCAGCATTATTATCAAAGAAGGCGACGTTGGAAGCATAGTTTATGTTATGGAAGATGGCGCTTTAGCAGAAATTTAA
Protein
MSHTHTKQSQMEHNYKLEKKYLEMKRMVYERTEELKRRDKIITLLENEIDEKDAQIRYLKNEIDKYRQVVKPLTRQIIDSHKNGIDTDEECFTMKTPGVENTRVYSLNEPRLKRTAISAEPLSSLTQEHELKLLKIPKSHRSRELIKTAILDNDFMKNLEMTQIREIVDCMYPVEYAAGSIIIKEGDVGSIVYVMEDGALAEI
Summary
Description
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors.
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Component of the Mediator complex, which is composed of MED1, MED4, MED6, MED7, MED8, MED9, MED10, MED11, MED12, MED13, MED13L, MED14, MED15, MED16, MED17, MED18, MED19, MED20, MED21, MED22, MED23, MED24, MED25, MED26, MED27, MED29, MED30, MED31, CCNC, CDK8 and CDC2L6/CDK11. The MED12, MED13, CCNC and CDK8 subunits form a distinct module termed the CDK8 module. Mediator containing the CDK8 module is less active than Mediator lacking this module in supporting transcriptional activation. Individual preparations of the Mediator complex lacking one or more distinct subunits have been variously termed ARC, CRSP, DRIP, PC2, SMCC and TRAP.
Similarity
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cGMP subfamily.
Belongs to the Mediator complex subunit 4 family.
Belongs to the Mediator complex subunit 4 family.
Uniprot
H9JH58
A0A2H1W9V6
A0A194QIC8
A0A194R7S6
A0A2A4ITY4
A0A212EK53
+ More
A0A0L7LCS9 A0A182RUS6 A0A182LN13 A0A182NPY3 A0A182Y953 Q5TNQ0 A0A182I751 A0A1B0CTA2 A0A182Q6A5 A0A182VU81 A0A182XDF2 A0A182LWB4 A0A182K2C8 A0A182V0F5 A0A1S4GZF7 A0A182TVI1 W5JAF4 A0A182F5P8 A0A182JMX8 A0A182P699 A0A1B6MN91 B0WEA0 A0A1B6H3B7 V5GAA9 E0VGN9 A0A084VYK8 A0A182HFC5 A0A195BR46 Q170R4 F4W6N2 E9J3W6 A0A088AEL9 C3PUN5 A0A195CLN6 U4U839 A0A2J7QHI8 A0A2J7QHL2 A0A1I8M0W3 A0A0L0BX72 A0A067QRI0 A0A1J1IK64 A0A195DIV3 A0A1J1II54 A0A286NL54 A0A2A3EII9 A0A154PSC4 C3PUN4 A0A034VA88 A0A0A1XJT6 A0A139WDC9 A0A232F2K2 A0A1S3KIL0 A0A2K5MRU0 A0A3Q0IQ78 A0A2Y9RTP1 A0A2Y9RLI9 Q32KM5 A0A2U3WLJ6 A0A3Q0CEB2 A0A1U8C3D8 F6YCF0 A0A1B0FRC2 A0A2R8YE50 A0A2R8YH74 A0A1U7U448 A0A1B0A618 A0A3B3QXZ2 B1ALS0 A0A2K6QTG9 A0A2R8Y507 L8I106 S7PYU2 D2GZ26 A0A161MSN1 A0A2U3YC82 A0A060VTS8 A0A2K6N0U1 A0A091CXW1 A0A1A9YSE5 A0A3B5KDT0 A0A1A9V0Y8 A0A151NZZ3 A0A151P0K9 A0A060W1F2 A0A1B0BC86 G1RMZ8 A0A3B3Z234 A0A3P9PHF9 A0A287D7T9 G7PDG2 A0A2K5D639 A0A2K5RSD6 A0A2R8P3E2
A0A0L7LCS9 A0A182RUS6 A0A182LN13 A0A182NPY3 A0A182Y953 Q5TNQ0 A0A182I751 A0A1B0CTA2 A0A182Q6A5 A0A182VU81 A0A182XDF2 A0A182LWB4 A0A182K2C8 A0A182V0F5 A0A1S4GZF7 A0A182TVI1 W5JAF4 A0A182F5P8 A0A182JMX8 A0A182P699 A0A1B6MN91 B0WEA0 A0A1B6H3B7 V5GAA9 E0VGN9 A0A084VYK8 A0A182HFC5 A0A195BR46 Q170R4 F4W6N2 E9J3W6 A0A088AEL9 C3PUN5 A0A195CLN6 U4U839 A0A2J7QHI8 A0A2J7QHL2 A0A1I8M0W3 A0A0L0BX72 A0A067QRI0 A0A1J1IK64 A0A195DIV3 A0A1J1II54 A0A286NL54 A0A2A3EII9 A0A154PSC4 C3PUN4 A0A034VA88 A0A0A1XJT6 A0A139WDC9 A0A232F2K2 A0A1S3KIL0 A0A2K5MRU0 A0A3Q0IQ78 A0A2Y9RTP1 A0A2Y9RLI9 Q32KM5 A0A2U3WLJ6 A0A3Q0CEB2 A0A1U8C3D8 F6YCF0 A0A1B0FRC2 A0A2R8YE50 A0A2R8YH74 A0A1U7U448 A0A1B0A618 A0A3B3QXZ2 B1ALS0 A0A2K6QTG9 A0A2R8Y507 L8I106 S7PYU2 D2GZ26 A0A161MSN1 A0A2U3YC82 A0A060VTS8 A0A2K6N0U1 A0A091CXW1 A0A1A9YSE5 A0A3B5KDT0 A0A1A9V0Y8 A0A151NZZ3 A0A151P0K9 A0A060W1F2 A0A1B0BC86 G1RMZ8 A0A3B3Z234 A0A3P9PHF9 A0A287D7T9 G7PDG2 A0A2K5D639 A0A2K5RSD6 A0A2R8P3E2
EC Number
2.7.11.12
Pubmed
19121390
26354079
22118469
26227816
20966253
25244985
+ More
12364791 20920257 23761445 20566863 24438588 26483478 17510324 21719571 21282665 19332792 23537049 25315136 26108605 24845553 25348373 25830018 18362917 19820115 28648823 26383154 20431018 15164054 29240929 21269460 25362486 22751099 20010809 24755649 21551351 22293439 22002653
12364791 20920257 23761445 20566863 24438588 26483478 17510324 21719571 21282665 19332792 23537049 25315136 26108605 24845553 25348373 25830018 18362917 19820115 28648823 26383154 20431018 15164054 29240929 21269460 25362486 22751099 20010809 24755649 21551351 22293439 22002653
EMBL
BABH01012935
ODYU01007254
SOQ49869.1
KQ458756
KPJ05272.1
KQ460597
+ More
KPJ13888.1 NWSH01006819 PCG63225.1 AGBW02014320 OWR41861.1 JTDY01001633 KOB73292.1 AAAB01008984 EAL38953.3 APCN01003364 APCN01003365 AJWK01027341 AJWK01027342 AJWK01027343 AJWK01027344 AXCN02002088 AXCM01001316 ADMH02002048 ETN59764.1 GEBQ01028033 GEBQ01027389 GEBQ01026875 GEBQ01002605 JAT11944.1 JAT12588.1 JAT13102.1 JAT37372.1 DS231906 EDS45394.1 GECZ01000603 JAS69166.1 GALX01001406 JAB67060.1 AAZO01002204 DS235150 EEB12545.1 ATLV01018376 KE525231 KFB43052.1 JXUM01037095 JXUM01037096 JXUM01037097 KQ561117 KXJ79656.1 KQ976419 KYM89358.1 CH477469 EAT40455.1 GL887707 EGI70108.1 GL768069 EFZ12418.1 EF999976 ABW22624.1 KQ977595 KYN01615.1 KB632064 ERL88498.1 NEVH01013973 PNF28055.1 PNF28056.1 JRES01001205 KNC24596.1 KK853043 KDR12116.1 CVRI01000054 CRK99932.1 KQ980824 KYN12439.1 CRK99933.1 KX809576 ASZ69797.1 KZ288230 PBC31613.1 KQ435124 KZC14773.1 EF999975 ABW22623.1 GAKP01020474 JAC38478.1 GBXI01013756 GBXI01003142 JAD00536.1 JAD11150.1 KQ971361 KYB25936.1 NNAY01001164 OXU24925.1 BC110022 AAI10023.1 AAMC01046206 AAMC01046207 AAMC01046208 AAMC01046209 AAMC01046210 AAMC01046211 AAMC01046212 CCAG010005704 AC022537 AC068062 AC069079 AC073584 AL157399 AL596105 AL607031 AL731537 KF455236 JH882335 ELR49868.1 KE163776 EPQ13767.1 GL192388 EFB20288.1 GEMB01000717 JAS02419.1 FR904300 CDQ58257.1 KN123966 KFO22695.1 AKHW03001467 KYO42378.1 KYO42379.1 FR904360 CDQ60846.1 JXJN01011892 ADFV01114579 ADFV01114580 ADFV01114581 ADFV01114582 ADFV01114583 ADFV01114584 ADFV01114585 ADFV01114586 ADFV01114587 ADFV01114588 AGTP01107148 AGTP01107149 AGTP01107150 AGTP01107151 AGTP01107152 AGTP01107153 AGTP01107154 AGTP01107155 AGTP01107156 AGTP01107157 CM001284 EHH64844.1
KPJ13888.1 NWSH01006819 PCG63225.1 AGBW02014320 OWR41861.1 JTDY01001633 KOB73292.1 AAAB01008984 EAL38953.3 APCN01003364 APCN01003365 AJWK01027341 AJWK01027342 AJWK01027343 AJWK01027344 AXCN02002088 AXCM01001316 ADMH02002048 ETN59764.1 GEBQ01028033 GEBQ01027389 GEBQ01026875 GEBQ01002605 JAT11944.1 JAT12588.1 JAT13102.1 JAT37372.1 DS231906 EDS45394.1 GECZ01000603 JAS69166.1 GALX01001406 JAB67060.1 AAZO01002204 DS235150 EEB12545.1 ATLV01018376 KE525231 KFB43052.1 JXUM01037095 JXUM01037096 JXUM01037097 KQ561117 KXJ79656.1 KQ976419 KYM89358.1 CH477469 EAT40455.1 GL887707 EGI70108.1 GL768069 EFZ12418.1 EF999976 ABW22624.1 KQ977595 KYN01615.1 KB632064 ERL88498.1 NEVH01013973 PNF28055.1 PNF28056.1 JRES01001205 KNC24596.1 KK853043 KDR12116.1 CVRI01000054 CRK99932.1 KQ980824 KYN12439.1 CRK99933.1 KX809576 ASZ69797.1 KZ288230 PBC31613.1 KQ435124 KZC14773.1 EF999975 ABW22623.1 GAKP01020474 JAC38478.1 GBXI01013756 GBXI01003142 JAD00536.1 JAD11150.1 KQ971361 KYB25936.1 NNAY01001164 OXU24925.1 BC110022 AAI10023.1 AAMC01046206 AAMC01046207 AAMC01046208 AAMC01046209 AAMC01046210 AAMC01046211 AAMC01046212 CCAG010005704 AC022537 AC068062 AC069079 AC073584 AL157399 AL596105 AL607031 AL731537 KF455236 JH882335 ELR49868.1 KE163776 EPQ13767.1 GL192388 EFB20288.1 GEMB01000717 JAS02419.1 FR904300 CDQ58257.1 KN123966 KFO22695.1 AKHW03001467 KYO42378.1 KYO42379.1 FR904360 CDQ60846.1 JXJN01011892 ADFV01114579 ADFV01114580 ADFV01114581 ADFV01114582 ADFV01114583 ADFV01114584 ADFV01114585 ADFV01114586 ADFV01114587 ADFV01114588 AGTP01107148 AGTP01107149 AGTP01107150 AGTP01107151 AGTP01107152 AGTP01107153 AGTP01107154 AGTP01107155 AGTP01107156 AGTP01107157 CM001284 EHH64844.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000075900 UP000075882 UP000075884 UP000076408 UP000007062 UP000075840 UP000092461 UP000075886 UP000075920 UP000076407 UP000075883 UP000075881 UP000075903 UP000075902 UP000000673 UP000069272 UP000075880 UP000075885 UP000002320 UP000009046 UP000030765 UP000069940 UP000249989 UP000078540 UP000008820 UP000007755 UP000005203 UP000078542 UP000030742 UP000235965 UP000095301 UP000037069 UP000027135 UP000183832 UP000078492 UP000242457 UP000076502 UP000007266 UP000215335 UP000085678 UP000233060 UP000079169 UP000248480 UP000245340 UP000189706 UP000008143 UP000092444 UP000005640 UP000189704 UP000092445 UP000261540 UP000233200 UP000245341 UP000193380 UP000233180 UP000028990 UP000092443 UP000005226 UP000078200 UP000050525 UP000092460 UP000001073 UP000261480 UP000242638 UP000005215 UP000009130 UP000233020 UP000233040 UP000008225
UP000075900 UP000075882 UP000075884 UP000076408 UP000007062 UP000075840 UP000092461 UP000075886 UP000075920 UP000076407 UP000075883 UP000075881 UP000075903 UP000075902 UP000000673 UP000069272 UP000075880 UP000075885 UP000002320 UP000009046 UP000030765 UP000069940 UP000249989 UP000078540 UP000008820 UP000007755 UP000005203 UP000078542 UP000030742 UP000235965 UP000095301 UP000037069 UP000027135 UP000183832 UP000078492 UP000242457 UP000076502 UP000007266 UP000215335 UP000085678 UP000233060 UP000079169 UP000248480 UP000245340 UP000189706 UP000008143 UP000092444 UP000005640 UP000189704 UP000092445 UP000261540 UP000233200 UP000245341 UP000193380 UP000233180 UP000028990 UP000092443 UP000005226 UP000078200 UP000050525 UP000092460 UP000001073 UP000261480 UP000242638 UP000005215 UP000009130 UP000233020 UP000233040 UP000008225
Interpro
IPR018488
cNMP-bd_CS
+ More
IPR000595 cNMP-bd_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR012198 cAMP_dep_PK_reg_su
IPR002374 cGMP_dep_kinase
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR035014 STKc_cGK
IPR011009 Kinase-like_dom_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR031831 PKcGMP_CC
IPR019258 Mediator_Med4
IPR000595 cNMP-bd_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR012198 cAMP_dep_PK_reg_su
IPR002374 cGMP_dep_kinase
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR035014 STKc_cGK
IPR011009 Kinase-like_dom_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR031831 PKcGMP_CC
IPR019258 Mediator_Med4
Gene 3D
ProteinModelPortal
H9JH58
A0A2H1W9V6
A0A194QIC8
A0A194R7S6
A0A2A4ITY4
A0A212EK53
+ More
A0A0L7LCS9 A0A182RUS6 A0A182LN13 A0A182NPY3 A0A182Y953 Q5TNQ0 A0A182I751 A0A1B0CTA2 A0A182Q6A5 A0A182VU81 A0A182XDF2 A0A182LWB4 A0A182K2C8 A0A182V0F5 A0A1S4GZF7 A0A182TVI1 W5JAF4 A0A182F5P8 A0A182JMX8 A0A182P699 A0A1B6MN91 B0WEA0 A0A1B6H3B7 V5GAA9 E0VGN9 A0A084VYK8 A0A182HFC5 A0A195BR46 Q170R4 F4W6N2 E9J3W6 A0A088AEL9 C3PUN5 A0A195CLN6 U4U839 A0A2J7QHI8 A0A2J7QHL2 A0A1I8M0W3 A0A0L0BX72 A0A067QRI0 A0A1J1IK64 A0A195DIV3 A0A1J1II54 A0A286NL54 A0A2A3EII9 A0A154PSC4 C3PUN4 A0A034VA88 A0A0A1XJT6 A0A139WDC9 A0A232F2K2 A0A1S3KIL0 A0A2K5MRU0 A0A3Q0IQ78 A0A2Y9RTP1 A0A2Y9RLI9 Q32KM5 A0A2U3WLJ6 A0A3Q0CEB2 A0A1U8C3D8 F6YCF0 A0A1B0FRC2 A0A2R8YE50 A0A2R8YH74 A0A1U7U448 A0A1B0A618 A0A3B3QXZ2 B1ALS0 A0A2K6QTG9 A0A2R8Y507 L8I106 S7PYU2 D2GZ26 A0A161MSN1 A0A2U3YC82 A0A060VTS8 A0A2K6N0U1 A0A091CXW1 A0A1A9YSE5 A0A3B5KDT0 A0A1A9V0Y8 A0A151NZZ3 A0A151P0K9 A0A060W1F2 A0A1B0BC86 G1RMZ8 A0A3B3Z234 A0A3P9PHF9 A0A287D7T9 G7PDG2 A0A2K5D639 A0A2K5RSD6 A0A2R8P3E2
A0A0L7LCS9 A0A182RUS6 A0A182LN13 A0A182NPY3 A0A182Y953 Q5TNQ0 A0A182I751 A0A1B0CTA2 A0A182Q6A5 A0A182VU81 A0A182XDF2 A0A182LWB4 A0A182K2C8 A0A182V0F5 A0A1S4GZF7 A0A182TVI1 W5JAF4 A0A182F5P8 A0A182JMX8 A0A182P699 A0A1B6MN91 B0WEA0 A0A1B6H3B7 V5GAA9 E0VGN9 A0A084VYK8 A0A182HFC5 A0A195BR46 Q170R4 F4W6N2 E9J3W6 A0A088AEL9 C3PUN5 A0A195CLN6 U4U839 A0A2J7QHI8 A0A2J7QHL2 A0A1I8M0W3 A0A0L0BX72 A0A067QRI0 A0A1J1IK64 A0A195DIV3 A0A1J1II54 A0A286NL54 A0A2A3EII9 A0A154PSC4 C3PUN4 A0A034VA88 A0A0A1XJT6 A0A139WDC9 A0A232F2K2 A0A1S3KIL0 A0A2K5MRU0 A0A3Q0IQ78 A0A2Y9RTP1 A0A2Y9RLI9 Q32KM5 A0A2U3WLJ6 A0A3Q0CEB2 A0A1U8C3D8 F6YCF0 A0A1B0FRC2 A0A2R8YE50 A0A2R8YH74 A0A1U7U448 A0A1B0A618 A0A3B3QXZ2 B1ALS0 A0A2K6QTG9 A0A2R8Y507 L8I106 S7PYU2 D2GZ26 A0A161MSN1 A0A2U3YC82 A0A060VTS8 A0A2K6N0U1 A0A091CXW1 A0A1A9YSE5 A0A3B5KDT0 A0A1A9V0Y8 A0A151NZZ3 A0A151P0K9 A0A060W1F2 A0A1B0BC86 G1RMZ8 A0A3B3Z234 A0A3P9PHF9 A0A287D7T9 G7PDG2 A0A2K5D639 A0A2K5RSD6 A0A2R8P3E2
PDB
3SHR
E-value=1.45236e-22,
Score=259
Ontologies
GO
Topology
Subcellular location
Nucleus
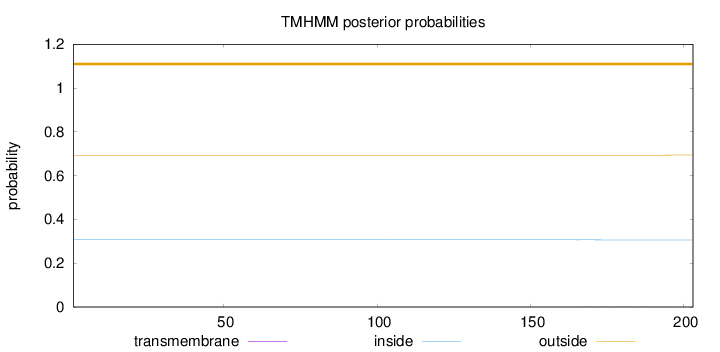
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01747
Exp number, first 60 AAs:
0
Total prob of N-in:
0.30690
outside
1 - 203
Population Genetic Test Statistics
Pi
144.756832
Theta
155.819621
Tajima's D
0.682891
CLR
0
CSRT
0.568071596420179
Interpretation
Uncertain
