Gene
KWMTBOMO01243
Pre Gene Modal
BGIBMGA008851
Annotation
PREDICTED:_dual_3'?5'-cyclic-AMP_and_-GMP_phosphodiesterase_11_isoform_X5_[Bombyx_mori]
Full name
Phosphodiesterase
+ More
Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11
Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11
Alternative Name
cAMP and cGMP phosphodiesterase 11
Location in the cell
Cytoplasmic Reliability : 1.425 Nuclear Reliability : 1.011 PlasmaMembrane Reliability : 1.45
Sequence
CDS
ATGACTGTGAACATCGCGGATGCTTATTTGGATGCCCGTTTTGATCCCGCCGTCGATGAAGGAACCGGCTTCAAGCACAATACGATCCTCTGCATGGCCATTAAGAATTCAGAGGGGAGGATAATCGGTGTTATTCAGTTGATCAACAAGTTTGATGACTTGTTGTTCACGAAGAACGATGAGAACTTCGTCGAAGCTTTTGCCATTTTTTGCGGAATGGGCATCCACAATACGCACATGTACGAGAAGGCGATCACGGCTATGGCGAAGCAAAGCGTCACGCTTGACGTGTTGAGCTACCACGCATCAGCTTCGTTGGACGATGCTCAGCGATTACGGTCCTTGTTCCAGTCGTTGAGGATACCGTCATCGGCTCACTTCAGCCTCCACGAGCTGGCGTTCGACGACATCAATATGACGGACGACGACACGTTGAGAGCGTGCCTCCGGATGTTTCTGGACATGGACTTTGTCGAGAGATTTCACATAGACTACGCGGTCTTGTGCCGGTGGCTGCTGAGCGTGAAGAAAAACTATCGGAACGTCACTTACCACAATTGGAGGCACGCTTTCAATGTGGCCCAAATGATGTTCGCAATATTGACGGAGACTCAGTGGTGGAAGATATTCGGCGAGCTGGAATGCTTAGCTTTAATTATTGGCTGTCTGTGTCACGACCTTGATCACCGAGGGACCAATAACTCTTTCCAAATAAAGGCGTCGTCGCCATTAGCGCAGCTGTATTCGACATCCACAATGGAGCATCATCACTTCGATCAGTGTCTGATGATCTTGAACTCTCCCGGGAACCAAATCCTGCTGAATCTGTCTTCAGACGATTACGAGAAGGTCGTCAAGGTACTCGAGGACGCTATACTCTCCACCGACTTGGCTGTTTATTTCAGCAAAAGGAAAGCCTTCATAGAGCTGGTGAACTCTGATCGGACGGTGACTCCGTTGTGGGGCGAGTGCGTGGAGAGGCGTGGGCTGCTACGCGCCATGCTCATGACGGCCTGTGACCTCGCCGCCATCACCAAGCCCTGGCCAATAGAGCGGCGCGTGGCAGGACTGGTCGCTGGAGAGTTCTTCAGACAGGGCGACCTGGAGAGACAGAACCTGAACTTGACCCCTATTGACATAATGAATAGAGACAAAGAGGACCAGCTTCCGGCCATGCAAGTCAAGTTCATTGATACGATCTGCCTTCCGATATACGAGGCATTCGCGCGTCTATCGCCGGCCCTGCAGCCGATGCTCGATCGCGTCGAGAGCAATCGCGCGCACTGGATCGAGATGTCGAACAACGACGAGAAAATCGATTACGGACTCGAACGGGAAGCGATCGATAAAAACGACAAGTCCAGCGTCAGCTCGCAGAATGTCGACGACACCGAGAATACGTACGAAGGGGACAGCAGCGGCGCCGTCAGTCTGTCCTCCGAGACTAACTCGAAGTATGAGAACCTCGAGAATCCGGTCGTGGGGCTGTTGTGTAACAACGCGCTCGCCATCCCGAGAGACGCGACCAAGTTCTGTCACATTAACGACGTCAACTAG
Protein
MTVNIADAYLDARFDPAVDEGTGFKHNTILCMAIKNSEGRIIGVIQLINKFDDLLFTKNDENFVEAFAIFCGMGIHNTHMYEKAITAMAKQSVTLDVLSYHASASLDDAQRLRSLFQSLRIPSSAHFSLHELAFDDINMTDDDTLRACLRMFLDMDFVERFHIDYAVLCRWLLSVKKNYRNVTYHNWRHAFNVAQMMFAILTETQWWKIFGELECLALIIGCLCHDLDHRGTNNSFQIKASSPLAQLYSTSTMEHHHFDQCLMILNSPGNQILLNLSSDDYEKVVKVLEDAILSTDLAVYFSKRKAFIELVNSDRTVTPLWGECVERRGLLRAMLMTACDLAAITKPWPIERRVAGLVAGEFFRQGDLERQNLNLTPIDIMNRDKEDQLPAMQVKFIDTICLPIYEAFARLSPALQPMLDRVESNRAHWIEMSNNDEKIDYGLEREAIDKNDKSSVSSQNVDDTENTYEGDSSGAVSLSSETNSKYENLENPVVGLLCNNALAIPRDATKFCHINDVN
Summary
Description
Plays a role in signal transduction by regulating the intracellular concentration of cyclic nucleotides cAMP and cGMP. Catalyzes the hydrolysis of both cAMP and cGMP to 5'-AMP and 5'-GMP, respectively.
Catalytic Activity
3',5'-cyclic GMP + H2O = GMP + H(+)
3',5'-cyclic AMP + H2O = AMP + H(+)
3',5'-cyclic AMP + H2O = AMP + H(+)
Cofactor
a divalent metal cation
Biophysicochemical Properties
18.5 uM for cAMP KM=6 uM for cGMP
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Keywords
Alternative splicing
cAMP
cGMP
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Repeat
Feature
chain Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A2W1BX00
A0A2A4JRT0
A0A2H1X2K2
H9JH54
A0A194R7T1
A0A1S4F7V7
+ More
Q17CK8 B0X978 A0A0L7RKM4 A0A1I8P1H4 A0A182G306 A0A195CCX4 A0A1B0FN80 A0A182GNV7 B4P9V0 A0A1A9VXM8 A0A1A9ZIV5 A0A0Q9WCQ4 B4LTJ1 B4KK28 B4JDM2 E1ZV78 A0A0Q9XE74 Q9VJ79-2 A0A1B0B4N4 E0VX88 B3MM25 A0A0Q5VZ46 A0A0J9R482 A0A026W6P7 Q9VJ79-3 A0A1Q3EX09 A0A195BFM4 X2J6P6 X2J8Z1 A0A0Q5VUH7 A0A0J9R528 Q9VJ79 T1PGJ0 A0A1Q3EW90 A0A1W4V9M4 A0A0Q9WBB0 A0A0Q9X413 A0A1A9XDR6 A0A1I8P1C0 A0A151WUW1 A0A1W4VMQ0 A0A088AHD5 A0A1B6F748 A0A0C9QUJ7 A0A154PAQ7 B4MZG4 A0A0A1XNM9 A0A0J7KKA5 A0A151JB35 M9MSJ0 A0A195FAQ6 A0A0R3NSE6 A0A1B6KEA3 A0A182KA65 A0A0Q5VKF0 A0A0J9R475 A0A084W7Q4 A0A1W4VM14 M9MRK1 W8AMK1 A0A0J9R3I8 A0A182J222 B4Q8U9 A0A0K8WEA3 B5DHZ2 B4I5H6 A0A0K8VLT8 A0A1I8MJ33 A0A0C9RZA0 E2BX21 A0A182NGQ3 A0A3B0J9Q2 A0A3B0JFK7 A0A0M4EE19 A0A067R4U9 A0A0L0CC81 A0A182TAU6 A0A0N0BHH4 A0A0R3NSI8 A0A1J1IHZ2 W8B7A9 D6WXS4 T1I6M0 A0A2M3Z0Y2 A0A2M4A1J8 U4U9I9 N6U5Q9 A0A2R7VRY1 A0A2M3Z0V7 A0A069DZL1 A0A2M3YYZ9 A0A1Y1MHF0 A0A182P1L3 A0A2M4BBA8
Q17CK8 B0X978 A0A0L7RKM4 A0A1I8P1H4 A0A182G306 A0A195CCX4 A0A1B0FN80 A0A182GNV7 B4P9V0 A0A1A9VXM8 A0A1A9ZIV5 A0A0Q9WCQ4 B4LTJ1 B4KK28 B4JDM2 E1ZV78 A0A0Q9XE74 Q9VJ79-2 A0A1B0B4N4 E0VX88 B3MM25 A0A0Q5VZ46 A0A0J9R482 A0A026W6P7 Q9VJ79-3 A0A1Q3EX09 A0A195BFM4 X2J6P6 X2J8Z1 A0A0Q5VUH7 A0A0J9R528 Q9VJ79 T1PGJ0 A0A1Q3EW90 A0A1W4V9M4 A0A0Q9WBB0 A0A0Q9X413 A0A1A9XDR6 A0A1I8P1C0 A0A151WUW1 A0A1W4VMQ0 A0A088AHD5 A0A1B6F748 A0A0C9QUJ7 A0A154PAQ7 B4MZG4 A0A0A1XNM9 A0A0J7KKA5 A0A151JB35 M9MSJ0 A0A195FAQ6 A0A0R3NSE6 A0A1B6KEA3 A0A182KA65 A0A0Q5VKF0 A0A0J9R475 A0A084W7Q4 A0A1W4VM14 M9MRK1 W8AMK1 A0A0J9R3I8 A0A182J222 B4Q8U9 A0A0K8WEA3 B5DHZ2 B4I5H6 A0A0K8VLT8 A0A1I8MJ33 A0A0C9RZA0 E2BX21 A0A182NGQ3 A0A3B0J9Q2 A0A3B0JFK7 A0A0M4EE19 A0A067R4U9 A0A0L0CC81 A0A182TAU6 A0A0N0BHH4 A0A0R3NSI8 A0A1J1IHZ2 W8B7A9 D6WXS4 T1I6M0 A0A2M3Z0Y2 A0A2M4A1J8 U4U9I9 N6U5Q9 A0A2R7VRY1 A0A2M3Z0V7 A0A069DZL1 A0A2M3YYZ9 A0A1Y1MHF0 A0A182P1L3 A0A2M4BBA8
EC Number
3.1.4.-
Pubmed
28756777
19121390
26354079
17510324
26483478
17994087
+ More
17550304 18057021 20798317 10731132 12537572 12537569 15673286 20566863 22936249 24508170 30249741 12537568 12537573 12537574 16110336 17569856 17569867 25830018 15632085 24438588 24495485 25315136 24845553 26108605 18362917 19820115 23537049 26334808 28004739
17550304 18057021 20798317 10731132 12537572 12537569 15673286 20566863 22936249 24508170 30249741 12537568 12537573 12537574 16110336 17569856 17569867 25830018 15632085 24438588 24495485 25315136 24845553 26108605 18362917 19820115 23537049 26334808 28004739
EMBL
KZ149910
PZC78154.1
NWSH01000794
PCG74173.1
ODYU01012511
SOQ58864.1
+ More
BABH01012950 KQ460597 KPJ13893.1 CH477307 EAT44089.1 DS232522 EDS43010.1 KQ414568 KOC71369.1 JXUM01006736 JXUM01006737 JXUM01006738 JXUM01006739 JXUM01006740 JXUM01006741 JXUM01006742 JXUM01006743 JXUM01006744 KQ560221 KXJ83750.1 KQ977935 KYM98570.1 CCAG010022043 JXUM01014948 JXUM01014949 JXUM01014950 KQ560417 KXJ82517.1 CM000158 EDW90291.2 CH940649 KRF81958.1 EDW64964.2 KRF81959.1 CH933807 EDW11546.2 CH916368 EDW03392.1 GL434441 EFN74937.1 KRG02750.1 AE014134 AY122262 JXJN01008376 DS235829 EEB17994.1 CH902620 EDV30840.2 CH954179 KQS61928.1 CM002910 KMY90831.1 KK107373 QOIP01000004 EZA51643.1 RLU23841.1 GFDL01015196 JAV19849.1 KQ976502 KYM82982.1 AHN54558.1 AHN54559.1 KQS61929.1 KMY90824.1 KA646973 AFP61602.1 GFDL01015471 JAV19574.1 KRF81960.1 KRG02752.1 KQ982730 KYQ51491.1 GECZ01023784 JAS45985.1 GBYB01004307 JAG74074.1 KQ434860 KZC08932.1 CH963920 EDW77749.2 GBXI01001735 JAD12557.1 LBMM01006380 KMQ90654.1 KQ979181 KYN22352.1 ADV37086.1 KQ981700 KYN37518.1 CH379060 KRT04088.1 GEBQ01030218 JAT09759.1 KQS61927.1 KMY90826.1 ATLV01021272 KE525315 KFB46248.1 ADV37087.1 GAMC01020837 GAMC01020834 JAB85718.1 KMY90827.1 CM000361 EDX05381.1 GDHF01003134 JAI49180.1 EDY70048.2 CH480822 EDW55632.1 GDHF01012467 GDHF01011566 JAI39847.1 JAI40748.1 GBYB01013436 JAG83203.1 GL451202 EFN79740.1 OUUW01000004 SPP79004.1 SPP79002.1 CP012523 ALC40197.1 KK852699 KDR18162.1 JRES01000613 KNC29856.1 KQ435754 KOX76072.1 KRT04087.1 CVRI01000054 CRK99879.1 GAMC01020831 JAB85724.1 KQ971362 EFA07940.2 ACPB03010138 GGFM01001422 MBW22173.1 GGFK01001365 MBW34686.1 KB632186 ERL89722.1 APGK01039019 APGK01039020 APGK01039021 APGK01039022 APGK01039023 KB740966 ENN76950.1 KK854049 PTY10277.1 GGFM01001416 MBW22167.1 GBGD01000450 JAC88439.1 GGFM01000734 MBW21485.1 GEZM01030960 JAV85134.1 GGFJ01001169 MBW50310.1
BABH01012950 KQ460597 KPJ13893.1 CH477307 EAT44089.1 DS232522 EDS43010.1 KQ414568 KOC71369.1 JXUM01006736 JXUM01006737 JXUM01006738 JXUM01006739 JXUM01006740 JXUM01006741 JXUM01006742 JXUM01006743 JXUM01006744 KQ560221 KXJ83750.1 KQ977935 KYM98570.1 CCAG010022043 JXUM01014948 JXUM01014949 JXUM01014950 KQ560417 KXJ82517.1 CM000158 EDW90291.2 CH940649 KRF81958.1 EDW64964.2 KRF81959.1 CH933807 EDW11546.2 CH916368 EDW03392.1 GL434441 EFN74937.1 KRG02750.1 AE014134 AY122262 JXJN01008376 DS235829 EEB17994.1 CH902620 EDV30840.2 CH954179 KQS61928.1 CM002910 KMY90831.1 KK107373 QOIP01000004 EZA51643.1 RLU23841.1 GFDL01015196 JAV19849.1 KQ976502 KYM82982.1 AHN54558.1 AHN54559.1 KQS61929.1 KMY90824.1 KA646973 AFP61602.1 GFDL01015471 JAV19574.1 KRF81960.1 KRG02752.1 KQ982730 KYQ51491.1 GECZ01023784 JAS45985.1 GBYB01004307 JAG74074.1 KQ434860 KZC08932.1 CH963920 EDW77749.2 GBXI01001735 JAD12557.1 LBMM01006380 KMQ90654.1 KQ979181 KYN22352.1 ADV37086.1 KQ981700 KYN37518.1 CH379060 KRT04088.1 GEBQ01030218 JAT09759.1 KQS61927.1 KMY90826.1 ATLV01021272 KE525315 KFB46248.1 ADV37087.1 GAMC01020837 GAMC01020834 JAB85718.1 KMY90827.1 CM000361 EDX05381.1 GDHF01003134 JAI49180.1 EDY70048.2 CH480822 EDW55632.1 GDHF01012467 GDHF01011566 JAI39847.1 JAI40748.1 GBYB01013436 JAG83203.1 GL451202 EFN79740.1 OUUW01000004 SPP79004.1 SPP79002.1 CP012523 ALC40197.1 KK852699 KDR18162.1 JRES01000613 KNC29856.1 KQ435754 KOX76072.1 KRT04087.1 CVRI01000054 CRK99879.1 GAMC01020831 JAB85724.1 KQ971362 EFA07940.2 ACPB03010138 GGFM01001422 MBW22173.1 GGFK01001365 MBW34686.1 KB632186 ERL89722.1 APGK01039019 APGK01039020 APGK01039021 APGK01039022 APGK01039023 KB740966 ENN76950.1 KK854049 PTY10277.1 GGFM01001416 MBW22167.1 GBGD01000450 JAC88439.1 GGFM01000734 MBW21485.1 GEZM01030960 JAV85134.1 GGFJ01001169 MBW50310.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000008820
UP000002320
UP000053825
+ More
UP000095300 UP000069940 UP000249989 UP000078542 UP000092444 UP000002282 UP000078200 UP000092445 UP000008792 UP000009192 UP000001070 UP000000311 UP000000803 UP000092460 UP000009046 UP000007801 UP000008711 UP000053097 UP000279307 UP000078540 UP000192221 UP000092443 UP000075809 UP000005203 UP000076502 UP000007798 UP000036403 UP000078492 UP000078541 UP000001819 UP000075881 UP000030765 UP000075880 UP000000304 UP000001292 UP000095301 UP000008237 UP000075884 UP000268350 UP000092553 UP000027135 UP000037069 UP000075901 UP000053105 UP000183832 UP000007266 UP000015103 UP000030742 UP000019118 UP000075885
UP000095300 UP000069940 UP000249989 UP000078542 UP000092444 UP000002282 UP000078200 UP000092445 UP000008792 UP000009192 UP000001070 UP000000311 UP000000803 UP000092460 UP000009046 UP000007801 UP000008711 UP000053097 UP000279307 UP000078540 UP000192221 UP000092443 UP000075809 UP000005203 UP000076502 UP000007798 UP000036403 UP000078492 UP000078541 UP000001819 UP000075881 UP000030765 UP000075880 UP000000304 UP000001292 UP000095301 UP000008237 UP000075884 UP000268350 UP000092553 UP000027135 UP000037069 UP000075901 UP000053105 UP000183832 UP000007266 UP000015103 UP000030742 UP000019118 UP000075885
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2W1BX00
A0A2A4JRT0
A0A2H1X2K2
H9JH54
A0A194R7T1
A0A1S4F7V7
+ More
Q17CK8 B0X978 A0A0L7RKM4 A0A1I8P1H4 A0A182G306 A0A195CCX4 A0A1B0FN80 A0A182GNV7 B4P9V0 A0A1A9VXM8 A0A1A9ZIV5 A0A0Q9WCQ4 B4LTJ1 B4KK28 B4JDM2 E1ZV78 A0A0Q9XE74 Q9VJ79-2 A0A1B0B4N4 E0VX88 B3MM25 A0A0Q5VZ46 A0A0J9R482 A0A026W6P7 Q9VJ79-3 A0A1Q3EX09 A0A195BFM4 X2J6P6 X2J8Z1 A0A0Q5VUH7 A0A0J9R528 Q9VJ79 T1PGJ0 A0A1Q3EW90 A0A1W4V9M4 A0A0Q9WBB0 A0A0Q9X413 A0A1A9XDR6 A0A1I8P1C0 A0A151WUW1 A0A1W4VMQ0 A0A088AHD5 A0A1B6F748 A0A0C9QUJ7 A0A154PAQ7 B4MZG4 A0A0A1XNM9 A0A0J7KKA5 A0A151JB35 M9MSJ0 A0A195FAQ6 A0A0R3NSE6 A0A1B6KEA3 A0A182KA65 A0A0Q5VKF0 A0A0J9R475 A0A084W7Q4 A0A1W4VM14 M9MRK1 W8AMK1 A0A0J9R3I8 A0A182J222 B4Q8U9 A0A0K8WEA3 B5DHZ2 B4I5H6 A0A0K8VLT8 A0A1I8MJ33 A0A0C9RZA0 E2BX21 A0A182NGQ3 A0A3B0J9Q2 A0A3B0JFK7 A0A0M4EE19 A0A067R4U9 A0A0L0CC81 A0A182TAU6 A0A0N0BHH4 A0A0R3NSI8 A0A1J1IHZ2 W8B7A9 D6WXS4 T1I6M0 A0A2M3Z0Y2 A0A2M4A1J8 U4U9I9 N6U5Q9 A0A2R7VRY1 A0A2M3Z0V7 A0A069DZL1 A0A2M3YYZ9 A0A1Y1MHF0 A0A182P1L3 A0A2M4BBA8
Q17CK8 B0X978 A0A0L7RKM4 A0A1I8P1H4 A0A182G306 A0A195CCX4 A0A1B0FN80 A0A182GNV7 B4P9V0 A0A1A9VXM8 A0A1A9ZIV5 A0A0Q9WCQ4 B4LTJ1 B4KK28 B4JDM2 E1ZV78 A0A0Q9XE74 Q9VJ79-2 A0A1B0B4N4 E0VX88 B3MM25 A0A0Q5VZ46 A0A0J9R482 A0A026W6P7 Q9VJ79-3 A0A1Q3EX09 A0A195BFM4 X2J6P6 X2J8Z1 A0A0Q5VUH7 A0A0J9R528 Q9VJ79 T1PGJ0 A0A1Q3EW90 A0A1W4V9M4 A0A0Q9WBB0 A0A0Q9X413 A0A1A9XDR6 A0A1I8P1C0 A0A151WUW1 A0A1W4VMQ0 A0A088AHD5 A0A1B6F748 A0A0C9QUJ7 A0A154PAQ7 B4MZG4 A0A0A1XNM9 A0A0J7KKA5 A0A151JB35 M9MSJ0 A0A195FAQ6 A0A0R3NSE6 A0A1B6KEA3 A0A182KA65 A0A0Q5VKF0 A0A0J9R475 A0A084W7Q4 A0A1W4VM14 M9MRK1 W8AMK1 A0A0J9R3I8 A0A182J222 B4Q8U9 A0A0K8WEA3 B5DHZ2 B4I5H6 A0A0K8VLT8 A0A1I8MJ33 A0A0C9RZA0 E2BX21 A0A182NGQ3 A0A3B0J9Q2 A0A3B0JFK7 A0A0M4EE19 A0A067R4U9 A0A0L0CC81 A0A182TAU6 A0A0N0BHH4 A0A0R3NSI8 A0A1J1IHZ2 W8B7A9 D6WXS4 T1I6M0 A0A2M3Z0Y2 A0A2M4A1J8 U4U9I9 N6U5Q9 A0A2R7VRY1 A0A2M3Z0V7 A0A069DZL1 A0A2M3YYZ9 A0A1Y1MHF0 A0A182P1L3 A0A2M4BBA8
PDB
3BJC
E-value=1.77073e-123,
Score=1134
Ontologies
PATHWAY
GO
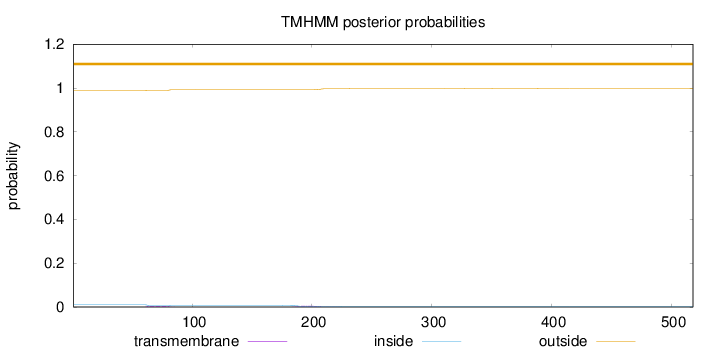
Topology
Length:
518
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18852
Exp number, first 60 AAs:
0.0011
Total prob of N-in:
0.01054
outside
1 - 518
Population Genetic Test Statistics
Pi
148.138446
Theta
179.863721
Tajima's D
-1.022427
CLR
1487.627695
CSRT
0.133793310334483
Interpretation
Uncertain
