Gene
KWMTBOMO01240
Pre Gene Modal
BGIBMGA008850
Annotation
PREDICTED:_dual_3'?5'-cyclic-AMP_and_-GMP_phosphodiesterase_11_isoform_X4_[Bombyx_mori]
Full name
Phosphodiesterase
Location in the cell
Nuclear Reliability : 2.158
Sequence
CDS
ATGGATTATTTGTTGTGCGATTACTGTGGTCTACATTTGCAGAGCATAGAAGAGCCGGTGGGTGGGGCGGGCGGTGGCATGCCCGCGCCCACCTGCTACGACGCGGAGTGTGCCCGCACGGAAGCCTGGCTCGATGAAAACCAGGAGTTCGTGCATGACTACTTCTTGAGGAAAGCGACGCGGCAAGTGGTGGACGCGTGGTTGGTTTCGCACGCGACCCCTCCCAGCGCTGAGCTAGCGTCGCCGTCGAGGGCTGGCTCTGGATCTGGAGCGACAACACCTGTCAGGAAGATCTCGGCGCATGAGTTCGAGCGCGGTGGTCTTTTGAAGCCGCTGGTAACCACCGTGGATGGCACGCCAACTTTCCTCGGAGACCAGCCGCCGCATGCCACTCCGACACGACCGCAGCGTCGCTCCAGGCATGAGCTGAGGCAGCTAGATGAGAAGGAGCTTATATTTGAGCTACGGTGA
Protein
MDYLLCDYCGLHLQSIEEPVGGAGGGMPAPTCYDAECARTEAWLDENQEFVHDYFLRKATRQVVDAWLVSHATPPSAELASPSRAGSGSGATTPVRKISAHEFERGGLLKPLVTTVDGTPTFLGDQPPHATPTRPQRRSRHELRQLDEKELIFELR
Summary
Cofactor
a divalent metal cation
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Uniprot
A0A212F5Z9
A0A2A4JRT0
A0A2W1BX00
A0A194R7T1
A0A0T6B596
D6WXS4
+ More
V5GVX2 N6U5Q9 U4U9I9 A0A067R4U9 A0A0J7NDR4 A0A026W6P7 A0A182G307 A0A336LU68 A0A195CCX4 A0A1B6EU68 A0A336LQJ2 A0A182X5V4 A0A336LTF2 A0A158NNF5 A0A1L8DLW0 A0A1B6KQ32 A0A1B6FST7 A0A182P1L5 A0A1B6MJU5 A0A182QHF9 A0A1B6IEG8 A0A151WUW1 A0A088AHD5 A0A2P8Z073 A0A2J7QW65 A0A0C9RZC1 A0A182MGZ5 A0A0L7RKM4 Q17CK8 E0VX88 E9IP50 A0A1S4F7V7 A0A1Y9IUW3 A0A182YN76 Q7Q8G2 A0A1I8JU78 E1ZV78 A0A0C9RZA0 A0A0C9QUJ7 A0A2C9GQ76 A0A2C9GQ13 A0A1B0DLY5 A0A154PAQ7 A0A2A3E526 A0A1Q3EX09 A0A2M4A1J8 A0A1Q3EW90 A0A182NGQ5 A0A3Q0IYF6 A0A2M4A2J7 A0A2M4A237 A0A2M4CII7 B0X978 A0A2M4A1Y8 A0A182F366 A0A2M4CSJ0 A0A2M3YZ59 A0A2M3YYZ9 A0A2M3Z0K9 A0A2M4BBA8 A0A2M4BBX5 A0A182J222 A0A1B0FN80 A0A1A9W0L3 A0A084W7Q4 E2BX21 A0A182V3S2 A0A1J1IHZ2 A0A2J7QW72 A0A2S2NQX7 T1JEP4
V5GVX2 N6U5Q9 U4U9I9 A0A067R4U9 A0A0J7NDR4 A0A026W6P7 A0A182G307 A0A336LU68 A0A195CCX4 A0A1B6EU68 A0A336LQJ2 A0A182X5V4 A0A336LTF2 A0A158NNF5 A0A1L8DLW0 A0A1B6KQ32 A0A1B6FST7 A0A182P1L5 A0A1B6MJU5 A0A182QHF9 A0A1B6IEG8 A0A151WUW1 A0A088AHD5 A0A2P8Z073 A0A2J7QW65 A0A0C9RZC1 A0A182MGZ5 A0A0L7RKM4 Q17CK8 E0VX88 E9IP50 A0A1S4F7V7 A0A1Y9IUW3 A0A182YN76 Q7Q8G2 A0A1I8JU78 E1ZV78 A0A0C9RZA0 A0A0C9QUJ7 A0A2C9GQ76 A0A2C9GQ13 A0A1B0DLY5 A0A154PAQ7 A0A2A3E526 A0A1Q3EX09 A0A2M4A1J8 A0A1Q3EW90 A0A182NGQ5 A0A3Q0IYF6 A0A2M4A2J7 A0A2M4A237 A0A2M4CII7 B0X978 A0A2M4A1Y8 A0A182F366 A0A2M4CSJ0 A0A2M3YZ59 A0A2M3YYZ9 A0A2M3Z0K9 A0A2M4BBA8 A0A2M4BBX5 A0A182J222 A0A1B0FN80 A0A1A9W0L3 A0A084W7Q4 E2BX21 A0A182V3S2 A0A1J1IHZ2 A0A2J7QW72 A0A2S2NQX7 T1JEP4
EC Number
3.1.4.-
Pubmed
EMBL
AGBW02010099
OWR49165.1
NWSH01000794
PCG74173.1
KZ149910
PZC78154.1
+ More
KQ460597 KPJ13893.1 LJIG01009690 KRT82576.1 KQ971362 EFA07940.2 GALX01004073 JAB64393.1 APGK01039019 APGK01039020 APGK01039021 APGK01039022 APGK01039023 KB740966 ENN76950.1 KB632186 ERL89722.1 KK852699 KDR18162.1 LBMM01006380 KMQ90655.1 KK107373 QOIP01000004 EZA51643.1 RLU23841.1 JXUM01006749 JXUM01006750 JXUM01006751 JXUM01006752 JXUM01006753 KQ560221 KXJ83751.1 UFQT01000114 SSX20233.1 KQ977935 KYM98570.1 GECZ01028280 JAS41489.1 SSX20230.1 SSX20231.1 ADTU01021376 ADTU01021377 ADTU01021378 ADTU01021379 ADTU01021380 ADTU01021381 ADTU01021382 ADTU01021383 ADTU01021384 ADTU01021385 GFDF01006727 JAV07357.1 GEBQ01026653 JAT13324.1 GECZ01016473 JAS53296.1 GEBQ01003804 JAT36173.1 AXCN02002079 GECU01022414 JAS85292.1 KQ982730 KYQ51491.1 PYGN01000258 PSN49905.1 NEVH01009767 PNF32827.1 GBYB01013471 JAG83238.1 AXCM01001804 KQ414568 KOC71369.1 CH477307 EAT44089.1 DS235829 EEB17994.1 GL764457 EFZ17651.1 AAAB01008944 EAA10146.3 GL434441 EFN74937.1 GBYB01013436 JAG83203.1 GBYB01004307 JAG74074.1 APCN01000703 APCN01000704 AJVK01016602 AJVK01016603 KQ434860 KZC08932.1 KZ288365 PBC26837.1 GFDL01015196 JAV19849.1 GGFK01001365 MBW34686.1 GFDL01015471 JAV19574.1 GGFK01001733 MBW35054.1 GGFK01001490 MBW34811.1 GGFL01000978 MBW65156.1 DS232522 EDS43010.1 GGFK01001482 MBW34803.1 GGFL01003963 MBW68141.1 GGFM01000796 MBW21547.1 GGFM01000734 MBW21485.1 GGFM01001316 MBW22067.1 GGFJ01001169 MBW50310.1 GGFJ01001405 MBW50546.1 CCAG010022043 ATLV01021272 KE525315 KFB46248.1 GL451202 EFN79740.1 CVRI01000054 CRK99879.1 PNF32826.1 GGMR01006966 MBY19585.1 JH432127
KQ460597 KPJ13893.1 LJIG01009690 KRT82576.1 KQ971362 EFA07940.2 GALX01004073 JAB64393.1 APGK01039019 APGK01039020 APGK01039021 APGK01039022 APGK01039023 KB740966 ENN76950.1 KB632186 ERL89722.1 KK852699 KDR18162.1 LBMM01006380 KMQ90655.1 KK107373 QOIP01000004 EZA51643.1 RLU23841.1 JXUM01006749 JXUM01006750 JXUM01006751 JXUM01006752 JXUM01006753 KQ560221 KXJ83751.1 UFQT01000114 SSX20233.1 KQ977935 KYM98570.1 GECZ01028280 JAS41489.1 SSX20230.1 SSX20231.1 ADTU01021376 ADTU01021377 ADTU01021378 ADTU01021379 ADTU01021380 ADTU01021381 ADTU01021382 ADTU01021383 ADTU01021384 ADTU01021385 GFDF01006727 JAV07357.1 GEBQ01026653 JAT13324.1 GECZ01016473 JAS53296.1 GEBQ01003804 JAT36173.1 AXCN02002079 GECU01022414 JAS85292.1 KQ982730 KYQ51491.1 PYGN01000258 PSN49905.1 NEVH01009767 PNF32827.1 GBYB01013471 JAG83238.1 AXCM01001804 KQ414568 KOC71369.1 CH477307 EAT44089.1 DS235829 EEB17994.1 GL764457 EFZ17651.1 AAAB01008944 EAA10146.3 GL434441 EFN74937.1 GBYB01013436 JAG83203.1 GBYB01004307 JAG74074.1 APCN01000703 APCN01000704 AJVK01016602 AJVK01016603 KQ434860 KZC08932.1 KZ288365 PBC26837.1 GFDL01015196 JAV19849.1 GGFK01001365 MBW34686.1 GFDL01015471 JAV19574.1 GGFK01001733 MBW35054.1 GGFK01001490 MBW34811.1 GGFL01000978 MBW65156.1 DS232522 EDS43010.1 GGFK01001482 MBW34803.1 GGFL01003963 MBW68141.1 GGFM01000796 MBW21547.1 GGFM01000734 MBW21485.1 GGFM01001316 MBW22067.1 GGFJ01001169 MBW50310.1 GGFJ01001405 MBW50546.1 CCAG010022043 ATLV01021272 KE525315 KFB46248.1 GL451202 EFN79740.1 CVRI01000054 CRK99879.1 PNF32826.1 GGMR01006966 MBY19585.1 JH432127
Proteomes
UP000007151
UP000218220
UP000053240
UP000007266
UP000019118
UP000030742
+ More
UP000027135 UP000036403 UP000053097 UP000279307 UP000069940 UP000249989 UP000078542 UP000076407 UP000005205 UP000075885 UP000075886 UP000075809 UP000005203 UP000245037 UP000235965 UP000075883 UP000053825 UP000008820 UP000009046 UP000075920 UP000076408 UP000007062 UP000075900 UP000000311 UP000075840 UP000092462 UP000076502 UP000242457 UP000075884 UP000079169 UP000002320 UP000069272 UP000075880 UP000092444 UP000091820 UP000030765 UP000008237 UP000075903 UP000183832
UP000027135 UP000036403 UP000053097 UP000279307 UP000069940 UP000249989 UP000078542 UP000076407 UP000005205 UP000075885 UP000075886 UP000075809 UP000005203 UP000245037 UP000235965 UP000075883 UP000053825 UP000008820 UP000009046 UP000075920 UP000076408 UP000007062 UP000075900 UP000000311 UP000075840 UP000092462 UP000076502 UP000242457 UP000075884 UP000079169 UP000002320 UP000069272 UP000075880 UP000092444 UP000091820 UP000030765 UP000008237 UP000075903 UP000183832
Interpro
SUPFAM
SSF46689
SSF46689
Gene 3D
CDD
ProteinModelPortal
A0A212F5Z9
A0A2A4JRT0
A0A2W1BX00
A0A194R7T1
A0A0T6B596
D6WXS4
+ More
V5GVX2 N6U5Q9 U4U9I9 A0A067R4U9 A0A0J7NDR4 A0A026W6P7 A0A182G307 A0A336LU68 A0A195CCX4 A0A1B6EU68 A0A336LQJ2 A0A182X5V4 A0A336LTF2 A0A158NNF5 A0A1L8DLW0 A0A1B6KQ32 A0A1B6FST7 A0A182P1L5 A0A1B6MJU5 A0A182QHF9 A0A1B6IEG8 A0A151WUW1 A0A088AHD5 A0A2P8Z073 A0A2J7QW65 A0A0C9RZC1 A0A182MGZ5 A0A0L7RKM4 Q17CK8 E0VX88 E9IP50 A0A1S4F7V7 A0A1Y9IUW3 A0A182YN76 Q7Q8G2 A0A1I8JU78 E1ZV78 A0A0C9RZA0 A0A0C9QUJ7 A0A2C9GQ76 A0A2C9GQ13 A0A1B0DLY5 A0A154PAQ7 A0A2A3E526 A0A1Q3EX09 A0A2M4A1J8 A0A1Q3EW90 A0A182NGQ5 A0A3Q0IYF6 A0A2M4A2J7 A0A2M4A237 A0A2M4CII7 B0X978 A0A2M4A1Y8 A0A182F366 A0A2M4CSJ0 A0A2M3YZ59 A0A2M3YYZ9 A0A2M3Z0K9 A0A2M4BBA8 A0A2M4BBX5 A0A182J222 A0A1B0FN80 A0A1A9W0L3 A0A084W7Q4 E2BX21 A0A182V3S2 A0A1J1IHZ2 A0A2J7QW72 A0A2S2NQX7 T1JEP4
V5GVX2 N6U5Q9 U4U9I9 A0A067R4U9 A0A0J7NDR4 A0A026W6P7 A0A182G307 A0A336LU68 A0A195CCX4 A0A1B6EU68 A0A336LQJ2 A0A182X5V4 A0A336LTF2 A0A158NNF5 A0A1L8DLW0 A0A1B6KQ32 A0A1B6FST7 A0A182P1L5 A0A1B6MJU5 A0A182QHF9 A0A1B6IEG8 A0A151WUW1 A0A088AHD5 A0A2P8Z073 A0A2J7QW65 A0A0C9RZC1 A0A182MGZ5 A0A0L7RKM4 Q17CK8 E0VX88 E9IP50 A0A1S4F7V7 A0A1Y9IUW3 A0A182YN76 Q7Q8G2 A0A1I8JU78 E1ZV78 A0A0C9RZA0 A0A0C9QUJ7 A0A2C9GQ76 A0A2C9GQ13 A0A1B0DLY5 A0A154PAQ7 A0A2A3E526 A0A1Q3EX09 A0A2M4A1J8 A0A1Q3EW90 A0A182NGQ5 A0A3Q0IYF6 A0A2M4A2J7 A0A2M4A237 A0A2M4CII7 B0X978 A0A2M4A1Y8 A0A182F366 A0A2M4CSJ0 A0A2M3YZ59 A0A2M3YYZ9 A0A2M3Z0K9 A0A2M4BBA8 A0A2M4BBX5 A0A182J222 A0A1B0FN80 A0A1A9W0L3 A0A084W7Q4 E2BX21 A0A182V3S2 A0A1J1IHZ2 A0A2J7QW72 A0A2S2NQX7 T1JEP4
PDB
3BJC
E-value=3.58441e-09,
Score=141
Ontologies
Topology
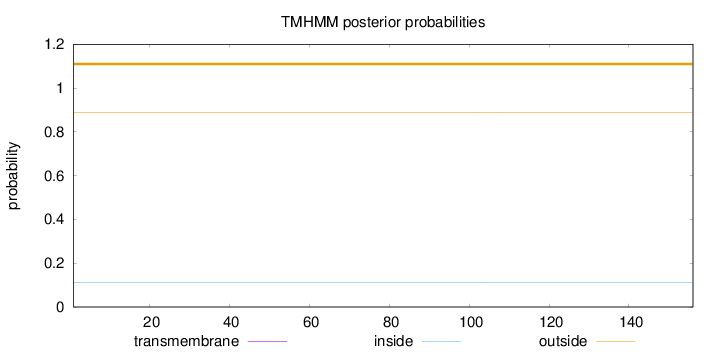
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.11048
outside
1 - 156
Population Genetic Test Statistics
Pi
247.862785
Theta
195.368141
Tajima's D
0.714135
CLR
0.535121
CSRT
0.575471226438678
Interpretation
Uncertain
