Gene
KWMTBOMO01236
Pre Gene Modal
BGIBMGA008849
Annotation
PREDICTED:_probable_G-protein_coupled_receptor_CG31760_[Amyelois_transitella]
Full name
Phosphodiesterase
+ More
Probable G-protein coupled receptor CG31760
Probable G-protein coupled receptor CG31760
Location in the cell
PlasmaMembrane Reliability : 4.03
Sequence
CDS
ATGGGGGGGCCGCGGCGCATGTCAAGTGCGCTGCTGCTGCCCTTCGCGTTGCTGTCGCTACCGCACACGAACGCCGCAAATGCAACCTCCTATCGGGAAGAGGTAGAATCATCATTGCGACTTGTGCACGCGGTGGCTACAGGAGCGACCGGCGCGCTTTGCGTGACACACCGCTACGGTCGTCTACGAGCACCGCTACACACGACACGATGGGACACGGCGCGGGCCCGAGCCGACTTGGCTGCCAATTTATTGCATCAGCTCGGATTGGAAACGGCTGAAGTACTCATGGCGGTTTCAAAAGGCTTAGTTTTGGGAGCCAGCCCGACCGAACGTGCTGTTGGATCGAGAGCATTAGCTCTGAGAAGCGATGGTGTTGTCAACAGTGCGGTCGCTTGGGAAAGATATGGAACCGCTGAGCCTGTTGCAGTTGTTTCACCGCAGTTGGAGAAGCCTCCGGACCCGGAATATCCATGGTATGTCTCAGCACACGAGAGCGAATCTCTTCGCTCGCCAAAGTTCATGCCTTCTTCTCCCGGTGCCACAATGGAAGGATGGTGGACTTATCCATACTACAGTTGCGGGGCGAAACGGTGGCTCTTATCTTATACCGTACCCGTCTCCACTATACGAGGGTTAAAAGGTATTGTTTCACTGGACATAGACATATCCAATTTGGAAATAAACCAGTGCGAGACAGATATATTTGATGAAAATGATAACCAAATATATTCCTTTCACGGAACTCATAAATGTCCCAATGAAACTACATACTGCGAATACAAACAGAGTCGTGAAATGAACGTTCGTCACACTGGTTGGGCGAGAGGATCATATGTTTGTAGATGCAGACCTGGCTTCTATTCAATTCATCATCCCGAAGGGTACAATGGATCTGTCATTGAAGTCGCTTACCAAGAATACGAAGAGAACGGATTAGAGAACTGGAGTGCACCATACGAATGCCTGAAATGTCAACCAGGATGTATTACATGCCGAGGCCCCGAACCATGCCTTGCCACCTACAACTGGCCATTTCGGATATCTTTGCTAGTAATATCAATGAGCTGTGTAGTGATGACTGTATGTCTTACAATATACACACGTCACCATCGCCGAGTAAAAGTGTTCCGAGTAGCCAGTCCTGTTTTTCTCTCAATTACTCTTCTTGGATGTGCCATTATGTACATGGAGATGGCTGCTATTTTCCCAGTTCTTGATAGGTACTCCTGTATTGCAACAAAATGGACTCGACACATGGGATTTTGTATTACATACACAGCACTTCTCATGAAGACTTGGAGAGTATCCCTCACATATCGGGTGAAATCAGCACACAAGTTGAAATTAACGGATAAACAATTATTGCAATGGATGGCTCCTATATTACTTATCATGTTGGTATACCTTTGTACATGGACATTGTCTTCACCGCCTGATGCCGAAGTCATAATGGACAATAAAGGATTGAAATTCAAGCAGTGTGTATATAATTGGTGGGATCATAGTTTAGCTATAGGTGAAATCCTTTTCCTGCTATGGGGTGTTCGTGTTTGTTACCGTGTACGTCACGCGGAGAGCTTGTACAATGAAGCTAAGCTAATCTCCTTTGCAATCTACAACATATTCACTGTAAATTCCTTGATGATTGCCTTCCATTTGCTGATATTACCACAAGCCGGTCCTGACATAAAATATTTGCTGGGATTCATAAGAACGCAACTGTCGACTTCGACTACGGTTTTGCTCGTATTTCTACCAAAAGTTTTACGCGTAGTTCGTGGTACGGGGGATACGTGGGATAGCAGGGCTCGCGCCCGAGGTGCTCCGGCTTCCTCATCGCTAAATGGCATCGGGCTGGTGCCAGACGAACCCCCGGATTTGTACCAGGAGAATGAAGAACTTAAGGAGGAAGTACAGAAACTGGCAGCCCAAATTGAATACATGAAAATAGTACAAATGGAGAAGAACAATCGGCATCTGAGACCGCGGCCCGGCGGCTACTTCACGTCCAATACCCAAGCGCCGCAGAGCCCCATGCACCCAAAGACCGTTTCTCAGGACAGTACAGAATACCCGGGGCCGGTGTGA
Protein
MGGPRRMSSALLLPFALLSLPHTNAANATSYREEVESSLRLVHAVATGATGALCVTHRYGRLRAPLHTTRWDTARARADLAANLLHQLGLETAEVLMAVSKGLVLGASPTERAVGSRALALRSDGVVNSAVAWERYGTAEPVAVVSPQLEKPPDPEYPWYVSAHESESLRSPKFMPSSPGATMEGWWTYPYYSCGAKRWLLSYTVPVSTIRGLKGIVSLDIDISNLEINQCETDIFDENDNQIYSFHGTHKCPNETTYCEYKQSREMNVRHTGWARGSYVCRCRPGFYSIHHPEGYNGSVIEVAYQEYEENGLENWSAPYECLKCQPGCITCRGPEPCLATYNWPFRISLLVISMSCVVMTVCLTIYTRHHRRVKVFRVASPVFLSITLLGCAIMYMEMAAIFPVLDRYSCIATKWTRHMGFCITYTALLMKTWRVSLTYRVKSAHKLKLTDKQLLQWMAPILLIMLVYLCTWTLSSPPDAEVIMDNKGLKFKQCVYNWWDHSLAIGEILFLLWGVRVCYRVRHAESLYNEAKLISFAIYNIFTVNSLMIAFHLLILPQAGPDIKYLLGFIRTQLSTSTTVLLVFLPKVLRVVRGTGDTWDSRARARGAPASSSLNGIGLVPDEPPDLYQENEELKEEVQKLAAQIEYMKIVQMEKNNRHLRPRPGGYFTSNTQAPQSPMHPKTVSQDSTEYPGPV
Summary
Cofactor
a divalent metal cation
Miscellaneous
This protein is translated by readthrough of a stop codon. Readthrough of the terminator codon TAG occurs between the codons for Val-875 and Arg-877. There is currently no sequence that provides the identity of residue 876.
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Belongs to the G-protein coupled receptor 3 family.
Belongs to the UDP-glycosyltransferase family.
Belongs to the G-protein coupled receptor 3 family.
Belongs to the UDP-glycosyltransferase family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Phosphoprotein
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Probable G-protein coupled receptor CG31760
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JH52
A0A2W1BWY9
A0A212ETU2
A0A0N1IPR9
A0A194QIQ3
A0A182NGQ0
+ More
A0A182Q216 A0A182UY53 A0A336LQJ2 A0A1Y9IVA0 A0A1S4GYU9 A0A336LTF2 A0A182TU96 A0A336LQD6 A0A0K8TZ62 A0A182YN74 A0A195CCP1 W8BYY8 A0A182RK45 A0A2C9GPG0 A0A151WU92 A0A182PVG6 A0A139WCX0 E1ZV76 A0A1B0FHI6 A0A1B0B2B2 A0A1W4WD73 A0A1W4W179 A0A1W4WVN1 Q9VKA4-2 B4Q3R4 B4P1Z6 A0A0J9R0U3 B3MNV6 A0A026W6G6 A0A3L8DV97 Q9VKA4 A0A1I8PLX4 Q7Q8G4 B3N3W2 A0A0L0BLI4 A0A0L7RKJ0 A0A1W4WUJ6 V9I8Z3 A0A1J1HE03 B4LTR5 B4JDG2 A0A3B0JG56 E2BX23 A0A1A9Y497 A0A2A3E6I8 T1I6W4 A0A088AHD6 B4KG94 V9IB91 A0A1W4WUM8 E0VX87 A0A151JPK2 F4W7Q7 A0A195BFP1 A0A0N0BHD2 B0X7M7 A0A158NNE5 Q29M70 A0A0Q9WZ96 A0A1B0AAM0 W5JC62 T1J8C6 Q17CL0 A0A2J7Q416 A0A182X5V8 A0A1A9WZK8 A0A182K2I9 A0A034WL08 A0A0N8E803 A0A0P5ZIS4 A0A0P6ENE5 A0A0P5CM02 A0A0P5ZVZ6 A0A0P4WP26 A0A0N8DQ29 A0A0P5CJ15 A0A0N8EIV7 A0A0P5HSN5 A0A0P4XGW0 A0A0P5HRP8 T1KRN3
A0A182Q216 A0A182UY53 A0A336LQJ2 A0A1Y9IVA0 A0A1S4GYU9 A0A336LTF2 A0A182TU96 A0A336LQD6 A0A0K8TZ62 A0A182YN74 A0A195CCP1 W8BYY8 A0A182RK45 A0A2C9GPG0 A0A151WU92 A0A182PVG6 A0A139WCX0 E1ZV76 A0A1B0FHI6 A0A1B0B2B2 A0A1W4WD73 A0A1W4W179 A0A1W4WVN1 Q9VKA4-2 B4Q3R4 B4P1Z6 A0A0J9R0U3 B3MNV6 A0A026W6G6 A0A3L8DV97 Q9VKA4 A0A1I8PLX4 Q7Q8G4 B3N3W2 A0A0L0BLI4 A0A0L7RKJ0 A0A1W4WUJ6 V9I8Z3 A0A1J1HE03 B4LTR5 B4JDG2 A0A3B0JG56 E2BX23 A0A1A9Y497 A0A2A3E6I8 T1I6W4 A0A088AHD6 B4KG94 V9IB91 A0A1W4WUM8 E0VX87 A0A151JPK2 F4W7Q7 A0A195BFP1 A0A0N0BHD2 B0X7M7 A0A158NNE5 Q29M70 A0A0Q9WZ96 A0A1B0AAM0 W5JC62 T1J8C6 Q17CL0 A0A2J7Q416 A0A182X5V8 A0A1A9WZK8 A0A182K2I9 A0A034WL08 A0A0N8E803 A0A0P5ZIS4 A0A0P6ENE5 A0A0P5CM02 A0A0P5ZVZ6 A0A0P4WP26 A0A0N8DQ29 A0A0P5CJ15 A0A0N8EIV7 A0A0P5HSN5 A0A0P4XGW0 A0A0P5HRP8 T1KRN3
EC Number
3.1.4.-
Pubmed
EMBL
BABH01012976
BABH01012977
KZ149910
PZC78144.1
AGBW02012537
OWR44871.1
+ More
KQ460317 KPJ16009.1 KQ458756 KPJ05279.1 AXCN02002080 UFQT01000114 SSX20230.1 AAAB01008944 SSX20231.1 SSX20232.1 GDHF01032587 JAI19727.1 KQ977935 KYM98572.1 GAMC01012123 GAMC01012121 JAB94434.1 APCN01000704 APCN01000705 APCN01000706 KQ982730 KYQ51489.1 KQ971362 KYB25725.1 GL434441 EFN74935.1 CCAG010008329 JXJN01007542 JXJN01007543 AE014134 AY118394 CM000361 EDX04789.1 CM000157 EDW88167.2 CM002910 KMY89887.1 CH902620 EDV32143.2 KK107373 EZA51640.1 QOIP01000004 RLU23839.1 EAA10329.3 CH954177 EDV58814.2 JRES01001697 KNC20882.1 KQ414568 KOC71375.1 JR037064 AEY57558.1 CVRI01000001 CRK86175.1 CH940649 EDW63966.2 CH916368 EDW03332.1 OUUW01000004 SPP79222.1 GL451202 EFN79742.1 KZ288365 PBC26836.1 ACPB03010138 CH933807 EDW11081.2 JR037065 AEY57559.1 DS235829 EEB17993.1 KQ978771 KYN28519.1 GL887862 EGI69763.1 KQ976502 KYM82985.1 KQ435754 KOX76068.1 DS232456 EDS42039.1 ADTU01021353 ADTU01021354 ADTU01021355 ADTU01021356 ADTU01021357 ADTU01021358 ADTU01021359 CH379060 EAL33824.3 CH963857 KRF98472.1 ADMH02001597 ETN61912.1 JH431954 CH477307 EAT44087.2 NEVH01018411 PNF23332.1 GAKP01002726 JAC56226.1 GDIQ01052844 JAN41893.1 GDIP01043846 JAM59869.1 GDIQ01073786 JAN20951.1 GDIP01168621 JAJ54781.1 GDIP01246060 GDIP01038986 JAM64729.1 GDIP01254613 JAI68788.1 GDIP01012015 JAM91700.1 GDIP01169808 JAJ53594.1 GDIQ01022412 JAN72325.1 GDIQ01223482 JAK28243.1 GDIP01241315 JAI82086.1 GDIQ01225888 JAK25837.1 CAEY01000414 CAEY01000415
KQ460317 KPJ16009.1 KQ458756 KPJ05279.1 AXCN02002080 UFQT01000114 SSX20230.1 AAAB01008944 SSX20231.1 SSX20232.1 GDHF01032587 JAI19727.1 KQ977935 KYM98572.1 GAMC01012123 GAMC01012121 JAB94434.1 APCN01000704 APCN01000705 APCN01000706 KQ982730 KYQ51489.1 KQ971362 KYB25725.1 GL434441 EFN74935.1 CCAG010008329 JXJN01007542 JXJN01007543 AE014134 AY118394 CM000361 EDX04789.1 CM000157 EDW88167.2 CM002910 KMY89887.1 CH902620 EDV32143.2 KK107373 EZA51640.1 QOIP01000004 RLU23839.1 EAA10329.3 CH954177 EDV58814.2 JRES01001697 KNC20882.1 KQ414568 KOC71375.1 JR037064 AEY57558.1 CVRI01000001 CRK86175.1 CH940649 EDW63966.2 CH916368 EDW03332.1 OUUW01000004 SPP79222.1 GL451202 EFN79742.1 KZ288365 PBC26836.1 ACPB03010138 CH933807 EDW11081.2 JR037065 AEY57559.1 DS235829 EEB17993.1 KQ978771 KYN28519.1 GL887862 EGI69763.1 KQ976502 KYM82985.1 KQ435754 KOX76068.1 DS232456 EDS42039.1 ADTU01021353 ADTU01021354 ADTU01021355 ADTU01021356 ADTU01021357 ADTU01021358 ADTU01021359 CH379060 EAL33824.3 CH963857 KRF98472.1 ADMH02001597 ETN61912.1 JH431954 CH477307 EAT44087.2 NEVH01018411 PNF23332.1 GAKP01002726 JAC56226.1 GDIQ01052844 JAN41893.1 GDIP01043846 JAM59869.1 GDIQ01073786 JAN20951.1 GDIP01168621 JAJ54781.1 GDIP01246060 GDIP01038986 JAM64729.1 GDIP01254613 JAI68788.1 GDIP01012015 JAM91700.1 GDIP01169808 JAJ53594.1 GDIQ01022412 JAN72325.1 GDIQ01223482 JAK28243.1 GDIP01241315 JAI82086.1 GDIQ01225888 JAK25837.1 CAEY01000414 CAEY01000415
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000075884
UP000075886
+ More
UP000075903 UP000075920 UP000075902 UP000076408 UP000078542 UP000075900 UP000075840 UP000075809 UP000075885 UP000007266 UP000000311 UP000092444 UP000092460 UP000192221 UP000192223 UP000000803 UP000000304 UP000002282 UP000007801 UP000053097 UP000279307 UP000095300 UP000007062 UP000008711 UP000037069 UP000053825 UP000183832 UP000008792 UP000001070 UP000268350 UP000008237 UP000092443 UP000242457 UP000015103 UP000005203 UP000009192 UP000009046 UP000078492 UP000007755 UP000078540 UP000053105 UP000002320 UP000005205 UP000001819 UP000007798 UP000092445 UP000000673 UP000008820 UP000235965 UP000076407 UP000091820 UP000075881 UP000015104
UP000075903 UP000075920 UP000075902 UP000076408 UP000078542 UP000075900 UP000075840 UP000075809 UP000075885 UP000007266 UP000000311 UP000092444 UP000092460 UP000192221 UP000192223 UP000000803 UP000000304 UP000002282 UP000007801 UP000053097 UP000279307 UP000095300 UP000007062 UP000008711 UP000037069 UP000053825 UP000183832 UP000008792 UP000001070 UP000268350 UP000008237 UP000092443 UP000242457 UP000015103 UP000005203 UP000009192 UP000009046 UP000078492 UP000007755 UP000078540 UP000053105 UP000002320 UP000005205 UP000001819 UP000007798 UP000092445 UP000000673 UP000008820 UP000235965 UP000076407 UP000091820 UP000075881 UP000015104
Pfam
Interpro
IPR017978
GPCR_3_C
+ More
IPR029016 GAF-like_dom_sf
IPR002073 PDEase_catalytic_dom
IPR036971 PDEase_catalytic_dom_sf
IPR003607 HD/PDEase_dom
IPR023174 PDEase_CS
IPR023088 PDEase
IPR003018 GAF
IPR009030 Growth_fac_rcpt_cys_sf
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR022533 Cox20
IPR036683 CO_DH_flav_C_dom_sf
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR000337 GPCR_3
IPR029016 GAF-like_dom_sf
IPR002073 PDEase_catalytic_dom
IPR036971 PDEase_catalytic_dom_sf
IPR003607 HD/PDEase_dom
IPR023174 PDEase_CS
IPR023088 PDEase
IPR003018 GAF
IPR009030 Growth_fac_rcpt_cys_sf
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR022533 Cox20
IPR036683 CO_DH_flav_C_dom_sf
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR000337 GPCR_3
SUPFAM
Gene 3D
ProteinModelPortal
H9JH52
A0A2W1BWY9
A0A212ETU2
A0A0N1IPR9
A0A194QIQ3
A0A182NGQ0
+ More
A0A182Q216 A0A182UY53 A0A336LQJ2 A0A1Y9IVA0 A0A1S4GYU9 A0A336LTF2 A0A182TU96 A0A336LQD6 A0A0K8TZ62 A0A182YN74 A0A195CCP1 W8BYY8 A0A182RK45 A0A2C9GPG0 A0A151WU92 A0A182PVG6 A0A139WCX0 E1ZV76 A0A1B0FHI6 A0A1B0B2B2 A0A1W4WD73 A0A1W4W179 A0A1W4WVN1 Q9VKA4-2 B4Q3R4 B4P1Z6 A0A0J9R0U3 B3MNV6 A0A026W6G6 A0A3L8DV97 Q9VKA4 A0A1I8PLX4 Q7Q8G4 B3N3W2 A0A0L0BLI4 A0A0L7RKJ0 A0A1W4WUJ6 V9I8Z3 A0A1J1HE03 B4LTR5 B4JDG2 A0A3B0JG56 E2BX23 A0A1A9Y497 A0A2A3E6I8 T1I6W4 A0A088AHD6 B4KG94 V9IB91 A0A1W4WUM8 E0VX87 A0A151JPK2 F4W7Q7 A0A195BFP1 A0A0N0BHD2 B0X7M7 A0A158NNE5 Q29M70 A0A0Q9WZ96 A0A1B0AAM0 W5JC62 T1J8C6 Q17CL0 A0A2J7Q416 A0A182X5V8 A0A1A9WZK8 A0A182K2I9 A0A034WL08 A0A0N8E803 A0A0P5ZIS4 A0A0P6ENE5 A0A0P5CM02 A0A0P5ZVZ6 A0A0P4WP26 A0A0N8DQ29 A0A0P5CJ15 A0A0N8EIV7 A0A0P5HSN5 A0A0P4XGW0 A0A0P5HRP8 T1KRN3
A0A182Q216 A0A182UY53 A0A336LQJ2 A0A1Y9IVA0 A0A1S4GYU9 A0A336LTF2 A0A182TU96 A0A336LQD6 A0A0K8TZ62 A0A182YN74 A0A195CCP1 W8BYY8 A0A182RK45 A0A2C9GPG0 A0A151WU92 A0A182PVG6 A0A139WCX0 E1ZV76 A0A1B0FHI6 A0A1B0B2B2 A0A1W4WD73 A0A1W4W179 A0A1W4WVN1 Q9VKA4-2 B4Q3R4 B4P1Z6 A0A0J9R0U3 B3MNV6 A0A026W6G6 A0A3L8DV97 Q9VKA4 A0A1I8PLX4 Q7Q8G4 B3N3W2 A0A0L0BLI4 A0A0L7RKJ0 A0A1W4WUJ6 V9I8Z3 A0A1J1HE03 B4LTR5 B4JDG2 A0A3B0JG56 E2BX23 A0A1A9Y497 A0A2A3E6I8 T1I6W4 A0A088AHD6 B4KG94 V9IB91 A0A1W4WUM8 E0VX87 A0A151JPK2 F4W7Q7 A0A195BFP1 A0A0N0BHD2 B0X7M7 A0A158NNE5 Q29M70 A0A0Q9WZ96 A0A1B0AAM0 W5JC62 T1J8C6 Q17CL0 A0A2J7Q416 A0A182X5V8 A0A1A9WZK8 A0A182K2I9 A0A034WL08 A0A0N8E803 A0A0P5ZIS4 A0A0P6ENE5 A0A0P5CM02 A0A0P5ZVZ6 A0A0P4WP26 A0A0N8DQ29 A0A0P5CJ15 A0A0N8EIV7 A0A0P5HSN5 A0A0P4XGW0 A0A0P5HRP8 T1KRN3
Ontologies
GO
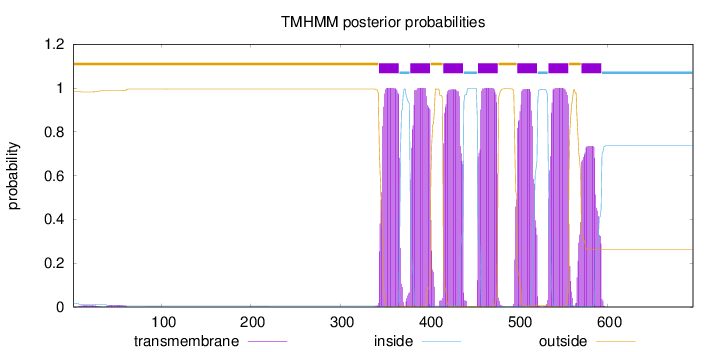
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 25,
Likelihood: 0.985641
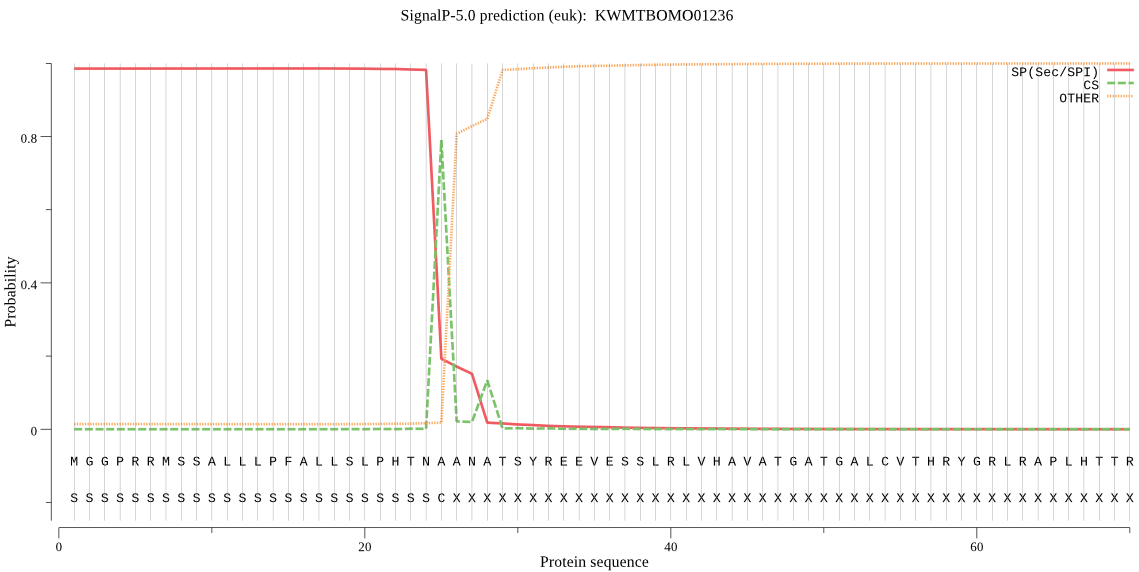
Length:
696
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
146.06239
Exp number, first 60 AAs:
0.32184
Total prob of N-in:
0.01680
outside
1 - 343
TMhelix
344 - 366
inside
367 - 378
TMhelix
379 - 401
outside
402 - 415
TMhelix
416 - 438
inside
439 - 454
TMhelix
455 - 477
outside
478 - 498
TMhelix
499 - 521
inside
522 - 533
TMhelix
534 - 556
outside
557 - 570
TMhelix
571 - 593
inside
594 - 696
Population Genetic Test Statistics
Pi
245.381009
Theta
197.763151
Tajima's D
0.81964
CLR
0.04733
CSRT
0.608469576521174
Interpretation
Uncertain
