Gene
KWMTBOMO01234
Annotation
Uncharacterized_protein_OBRU01_14001_[Operophtera_brumata]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 3.814
Sequence
CDS
ATGTATGTAAAACTAATGTTTGTTTCCCTTTTATGTACCTTTCAAACAGTTGTTTCGATAAAATGCTACCAATGCGCGTCTTCTCAAGATCACAAAGGGGAAGATACATGCGGAGCATACAGGAAATTCGATAGAGAAAGTCACATAGCTGTAGACTGTGGTAGCGATGAATCTCACATGCCAGGAGCATTTTGTATGAAGTTAACTCAACAAGGACCCAAAGGGTTCATTTGGGATGGCAGATGGCGTCAAGTTATAAGACGATGCGCATCTGTAGCTGAGACTGGAGTGACCGGTGTCTGTAACTGGGGTGTCTATGACAACGGTGTGTACTGGGAAGAATGTTACTGTACCTCAGATGAATGCAATGGTTCTACGAGTGTATCTCTCTCAATTGCATTATTCATGGGCTCATTGAGCATATTGTTTGGCTTGTATAAGGTCTAG
Protein
MYVKLMFVSLLCTFQTVVSIKCYQCASSQDHKGEDTCGAYRKFDRESHIAVDCGSDESHMPGAFCMKLTQQGPKGFIWDGRWRQVIRRCASVAETGVTGVCNWGVYDNGVYWEECYCTSDECNGSTSVSLSIALFMGSLSILFGLYKV
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Belongs to the AAA ATPase family.
Belongs to the AAA ATPase family.
Feature
chain Protein quiver
Uniprot
A0A2A4JKV3
A0A0L7L7B0
A0A2H1WRX4
A0A212EVR2
S4NXK5
A0A067RGD8
+ More
A0A1W4WK12 A0A0L0BT11 D6WXS0 A0A0A1WWD6 A0A0A1XHA5 A0A1B0CB01 A0A0T6B460 B3MNV4 A0A1Y1LY69 W8BKT3 B4KG96 A0A0K8WHA7 A0A2J7QW61 A0A1I8N3Q3 A0A1B6HNU8 A0A1L8DNY4 B3N3W0 B4LTR7 A0A0M4E7V2 Q9VKA1 B4IE77 A0A1B6L8F0 A0A1W4V309 B4P1Z9 Q8SZ82 Q29M71 B4G9N1 B4Q3R6 A0A3B0K6U2 A0A1J1HDY4 A0A1B0D4T5 Q71D51 A0A3B0JZU9 B4MVF1 B4JDG0 A0A1L8DNU6 A0A336LCK0 A0A182G4H0 A0A182YN73 A0A336LVK1 A0A1B6F5U8 A0A1A9VA77 A0A034WKB7 A0A182SY11 A0A023EH01 A0A1A9WZL2 Q17CL1 U5ESH5 A0A023EFJ7 A0A023EFB9 K7J3F8 A0A182IBJ2 A0A182VF76 A0A182NGP9 A0A1I8PYI1 A0A182RK47 A0A1S4GZK7 A0A1B0B2B4 A0A182WFF6 A0A1A9Y495 A0A182PVG5 E2B2C5 A0A1B0FHH4 E9FYS0 A0A1B0AAM8 A0A182KN54 E0VU09 A0A182JZ18 A0A182QCR2 W5JF11 A0A2M4C2G4 A0A182X5V9 A0A0A9XZ32 A0A087ZQE3 A0A2M4AVJ8 A0A182F363 A0A0C9QAZ2 A0A2A3E5P6 Q7Q8G5 A0A1D2MG56 A0A2M4C2D1 A0A1B6C834 A0A2M3ZED3 A0A182TXA0 A0A164Y888 A0A0P6ALF7 A0A0N8BIC2 A0A2M4C366 A0A182ILI7 A0A0P5VCG8 A0A1Q3F8D5 A0A2M4AYW3 A0A3L8DM28 A0A026W1W7
A0A1W4WK12 A0A0L0BT11 D6WXS0 A0A0A1WWD6 A0A0A1XHA5 A0A1B0CB01 A0A0T6B460 B3MNV4 A0A1Y1LY69 W8BKT3 B4KG96 A0A0K8WHA7 A0A2J7QW61 A0A1I8N3Q3 A0A1B6HNU8 A0A1L8DNY4 B3N3W0 B4LTR7 A0A0M4E7V2 Q9VKA1 B4IE77 A0A1B6L8F0 A0A1W4V309 B4P1Z9 Q8SZ82 Q29M71 B4G9N1 B4Q3R6 A0A3B0K6U2 A0A1J1HDY4 A0A1B0D4T5 Q71D51 A0A3B0JZU9 B4MVF1 B4JDG0 A0A1L8DNU6 A0A336LCK0 A0A182G4H0 A0A182YN73 A0A336LVK1 A0A1B6F5U8 A0A1A9VA77 A0A034WKB7 A0A182SY11 A0A023EH01 A0A1A9WZL2 Q17CL1 U5ESH5 A0A023EFJ7 A0A023EFB9 K7J3F8 A0A182IBJ2 A0A182VF76 A0A182NGP9 A0A1I8PYI1 A0A182RK47 A0A1S4GZK7 A0A1B0B2B4 A0A182WFF6 A0A1A9Y495 A0A182PVG5 E2B2C5 A0A1B0FHH4 E9FYS0 A0A1B0AAM8 A0A182KN54 E0VU09 A0A182JZ18 A0A182QCR2 W5JF11 A0A2M4C2G4 A0A182X5V9 A0A0A9XZ32 A0A087ZQE3 A0A2M4AVJ8 A0A182F363 A0A0C9QAZ2 A0A2A3E5P6 Q7Q8G5 A0A1D2MG56 A0A2M4C2D1 A0A1B6C834 A0A2M3ZED3 A0A182TXA0 A0A164Y888 A0A0P6ALF7 A0A0N8BIC2 A0A2M4C366 A0A182ILI7 A0A0P5VCG8 A0A1Q3F8D5 A0A2M4AYW3 A0A3L8DM28 A0A026W1W7
Pubmed
26227816
22118469
23622113
24845553
26108605
18362917
+ More
19820115 25830018 17994087 28004739 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 14525923 26483478 25244985 25348373 24945155 17510324 20075255 12364791 20798317 21292972 20966253 20566863 20920257 23761445 25401762 27289101 30249741 24508170
19820115 25830018 17994087 28004739 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 14525923 26483478 25244985 25348373 24945155 17510324 20075255 12364791 20798317 21292972 20966253 20566863 20920257 23761445 25401762 27289101 30249741 24508170
EMBL
NWSH01001143
PCG72416.1
JTDY01002449
KOB71392.1
ODYU01010604
SOQ55825.1
+ More
AGBW02012162 OWR45551.1 GAIX01012122 JAA80438.1 KK852699 KDR18169.1 JRES01001399 KNC23182.1 KQ971362 EFA07943.2 GBXI01011497 GBXI01011306 JAD02795.1 JAD02986.1 GBXI01003523 JAD10769.1 AJWK01004691 LJIG01016081 KRT81671.1 CH902620 EDV32141.2 GEZM01046792 GEZM01046791 JAV77005.1 GAMC01007173 JAB99382.1 CH933807 EDW11083.2 GDHF01001865 JAI50449.1 NEVH01009767 PNF32828.1 GECU01031349 JAS76357.1 GFDF01005925 JAV08159.1 CH954177 EDV58812.1 CH940649 EDW63968.1 CP012525 ALC42854.1 AE014134 BT056238 KX531468 AAF53177.1 ACL68685.1 ANY27278.1 CH480831 EDW45904.1 GEBQ01019985 JAT19992.1 CM000157 EDW88170.1 AY071055 AAL48677.1 CH379060 EAL33823.2 CH479180 EDW29061.1 CM000361 CM002910 EDX04791.1 KMY89894.1 OUUW01000004 SPP79218.1 CVRI01000001 CRK86176.1 AJVK01024863 AF532019 AAQ09918.1 SPP79219.1 CH963857 EDW76496.1 CH916368 EDW03330.1 GFDF01005955 JAV08129.1 UFQS01003100 UFQT01003100 SSX15199.1 SSX34574.1 JXUM01042352 KQ561321 KXJ78985.1 UFQT01000223 SSX21984.1 GECZ01024133 GECZ01022626 GECZ01012570 JAS45636.1 JAS47143.1 JAS57199.1 GAKP01004165 JAC54787.1 GAPW01005494 JAC08104.1 CH477307 EAT44086.1 GANO01002351 JAB57520.1 GAPW01005496 JAC08102.1 GAPW01005495 JAC08103.1 APCN01000707 AAAB01008944 JXJN01007544 GL445130 EFN90162.1 CCAG010008329 GL732527 EFX87583.1 DS235777 EEB16865.1 AXCN02002080 ADMH02001597 ETN61913.1 GGFJ01010308 MBW59449.1 GBHO01018430 GBRD01012933 JAG25174.1 JAG52893.1 GGFK01011492 MBW44813.1 GBYB01000399 JAG70166.1 KZ288389 PBC26391.1 EAA10154.4 LJIJ01001394 ODM91862.1 GGFJ01010303 MBW59444.1 GEDC01027704 JAS09594.1 GGFM01006121 MBW26872.1 LRGB01000930 KZS14980.1 GDIP01029307 JAM74408.1 GDIQ01174883 JAK76842.1 GGFJ01010307 MBW59448.1 GDIP01117065 JAL86649.1 GFDL01011214 JAV23831.1 GGFK01012658 MBW45979.1 QOIP01000006 RLU21484.1 KK107487 EZA50060.1
AGBW02012162 OWR45551.1 GAIX01012122 JAA80438.1 KK852699 KDR18169.1 JRES01001399 KNC23182.1 KQ971362 EFA07943.2 GBXI01011497 GBXI01011306 JAD02795.1 JAD02986.1 GBXI01003523 JAD10769.1 AJWK01004691 LJIG01016081 KRT81671.1 CH902620 EDV32141.2 GEZM01046792 GEZM01046791 JAV77005.1 GAMC01007173 JAB99382.1 CH933807 EDW11083.2 GDHF01001865 JAI50449.1 NEVH01009767 PNF32828.1 GECU01031349 JAS76357.1 GFDF01005925 JAV08159.1 CH954177 EDV58812.1 CH940649 EDW63968.1 CP012525 ALC42854.1 AE014134 BT056238 KX531468 AAF53177.1 ACL68685.1 ANY27278.1 CH480831 EDW45904.1 GEBQ01019985 JAT19992.1 CM000157 EDW88170.1 AY071055 AAL48677.1 CH379060 EAL33823.2 CH479180 EDW29061.1 CM000361 CM002910 EDX04791.1 KMY89894.1 OUUW01000004 SPP79218.1 CVRI01000001 CRK86176.1 AJVK01024863 AF532019 AAQ09918.1 SPP79219.1 CH963857 EDW76496.1 CH916368 EDW03330.1 GFDF01005955 JAV08129.1 UFQS01003100 UFQT01003100 SSX15199.1 SSX34574.1 JXUM01042352 KQ561321 KXJ78985.1 UFQT01000223 SSX21984.1 GECZ01024133 GECZ01022626 GECZ01012570 JAS45636.1 JAS47143.1 JAS57199.1 GAKP01004165 JAC54787.1 GAPW01005494 JAC08104.1 CH477307 EAT44086.1 GANO01002351 JAB57520.1 GAPW01005496 JAC08102.1 GAPW01005495 JAC08103.1 APCN01000707 AAAB01008944 JXJN01007544 GL445130 EFN90162.1 CCAG010008329 GL732527 EFX87583.1 DS235777 EEB16865.1 AXCN02002080 ADMH02001597 ETN61913.1 GGFJ01010308 MBW59449.1 GBHO01018430 GBRD01012933 JAG25174.1 JAG52893.1 GGFK01011492 MBW44813.1 GBYB01000399 JAG70166.1 KZ288389 PBC26391.1 EAA10154.4 LJIJ01001394 ODM91862.1 GGFJ01010303 MBW59444.1 GEDC01027704 JAS09594.1 GGFM01006121 MBW26872.1 LRGB01000930 KZS14980.1 GDIP01029307 JAM74408.1 GDIQ01174883 JAK76842.1 GGFJ01010307 MBW59448.1 GDIP01117065 JAL86649.1 GFDL01011214 JAV23831.1 GGFK01012658 MBW45979.1 QOIP01000006 RLU21484.1 KK107487 EZA50060.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000027135
UP000192223
UP000037069
+ More
UP000007266 UP000092461 UP000007801 UP000009192 UP000235965 UP000095301 UP000008711 UP000008792 UP000092553 UP000000803 UP000001292 UP000192221 UP000002282 UP000001819 UP000008744 UP000000304 UP000268350 UP000183832 UP000092462 UP000007798 UP000001070 UP000069940 UP000249989 UP000076408 UP000078200 UP000075901 UP000091820 UP000008820 UP000002358 UP000075840 UP000075903 UP000075884 UP000095300 UP000075900 UP000092460 UP000075920 UP000092443 UP000075885 UP000008237 UP000092444 UP000000305 UP000092445 UP000075882 UP000009046 UP000075881 UP000075886 UP000000673 UP000076407 UP000005203 UP000069272 UP000242457 UP000007062 UP000094527 UP000075902 UP000076858 UP000075880 UP000279307 UP000053097
UP000007266 UP000092461 UP000007801 UP000009192 UP000235965 UP000095301 UP000008711 UP000008792 UP000092553 UP000000803 UP000001292 UP000192221 UP000002282 UP000001819 UP000008744 UP000000304 UP000268350 UP000183832 UP000092462 UP000007798 UP000001070 UP000069940 UP000249989 UP000076408 UP000078200 UP000075901 UP000091820 UP000008820 UP000002358 UP000075840 UP000075903 UP000075884 UP000095300 UP000075900 UP000092460 UP000075920 UP000092443 UP000075885 UP000008237 UP000092444 UP000000305 UP000092445 UP000075882 UP000009046 UP000075881 UP000075886 UP000000673 UP000076407 UP000005203 UP000069272 UP000242457 UP000007062 UP000094527 UP000075902 UP000076858 UP000075880 UP000279307 UP000053097
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JKV3
A0A0L7L7B0
A0A2H1WRX4
A0A212EVR2
S4NXK5
A0A067RGD8
+ More
A0A1W4WK12 A0A0L0BT11 D6WXS0 A0A0A1WWD6 A0A0A1XHA5 A0A1B0CB01 A0A0T6B460 B3MNV4 A0A1Y1LY69 W8BKT3 B4KG96 A0A0K8WHA7 A0A2J7QW61 A0A1I8N3Q3 A0A1B6HNU8 A0A1L8DNY4 B3N3W0 B4LTR7 A0A0M4E7V2 Q9VKA1 B4IE77 A0A1B6L8F0 A0A1W4V309 B4P1Z9 Q8SZ82 Q29M71 B4G9N1 B4Q3R6 A0A3B0K6U2 A0A1J1HDY4 A0A1B0D4T5 Q71D51 A0A3B0JZU9 B4MVF1 B4JDG0 A0A1L8DNU6 A0A336LCK0 A0A182G4H0 A0A182YN73 A0A336LVK1 A0A1B6F5U8 A0A1A9VA77 A0A034WKB7 A0A182SY11 A0A023EH01 A0A1A9WZL2 Q17CL1 U5ESH5 A0A023EFJ7 A0A023EFB9 K7J3F8 A0A182IBJ2 A0A182VF76 A0A182NGP9 A0A1I8PYI1 A0A182RK47 A0A1S4GZK7 A0A1B0B2B4 A0A182WFF6 A0A1A9Y495 A0A182PVG5 E2B2C5 A0A1B0FHH4 E9FYS0 A0A1B0AAM8 A0A182KN54 E0VU09 A0A182JZ18 A0A182QCR2 W5JF11 A0A2M4C2G4 A0A182X5V9 A0A0A9XZ32 A0A087ZQE3 A0A2M4AVJ8 A0A182F363 A0A0C9QAZ2 A0A2A3E5P6 Q7Q8G5 A0A1D2MG56 A0A2M4C2D1 A0A1B6C834 A0A2M3ZED3 A0A182TXA0 A0A164Y888 A0A0P6ALF7 A0A0N8BIC2 A0A2M4C366 A0A182ILI7 A0A0P5VCG8 A0A1Q3F8D5 A0A2M4AYW3 A0A3L8DM28 A0A026W1W7
A0A1W4WK12 A0A0L0BT11 D6WXS0 A0A0A1WWD6 A0A0A1XHA5 A0A1B0CB01 A0A0T6B460 B3MNV4 A0A1Y1LY69 W8BKT3 B4KG96 A0A0K8WHA7 A0A2J7QW61 A0A1I8N3Q3 A0A1B6HNU8 A0A1L8DNY4 B3N3W0 B4LTR7 A0A0M4E7V2 Q9VKA1 B4IE77 A0A1B6L8F0 A0A1W4V309 B4P1Z9 Q8SZ82 Q29M71 B4G9N1 B4Q3R6 A0A3B0K6U2 A0A1J1HDY4 A0A1B0D4T5 Q71D51 A0A3B0JZU9 B4MVF1 B4JDG0 A0A1L8DNU6 A0A336LCK0 A0A182G4H0 A0A182YN73 A0A336LVK1 A0A1B6F5U8 A0A1A9VA77 A0A034WKB7 A0A182SY11 A0A023EH01 A0A1A9WZL2 Q17CL1 U5ESH5 A0A023EFJ7 A0A023EFB9 K7J3F8 A0A182IBJ2 A0A182VF76 A0A182NGP9 A0A1I8PYI1 A0A182RK47 A0A1S4GZK7 A0A1B0B2B4 A0A182WFF6 A0A1A9Y495 A0A182PVG5 E2B2C5 A0A1B0FHH4 E9FYS0 A0A1B0AAM8 A0A182KN54 E0VU09 A0A182JZ18 A0A182QCR2 W5JF11 A0A2M4C2G4 A0A182X5V9 A0A0A9XZ32 A0A087ZQE3 A0A2M4AVJ8 A0A182F363 A0A0C9QAZ2 A0A2A3E5P6 Q7Q8G5 A0A1D2MG56 A0A2M4C2D1 A0A1B6C834 A0A2M3ZED3 A0A182TXA0 A0A164Y888 A0A0P6ALF7 A0A0N8BIC2 A0A2M4C366 A0A182ILI7 A0A0P5VCG8 A0A1Q3F8D5 A0A2M4AYW3 A0A3L8DM28 A0A026W1W7
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 19,
Likelihood: 0.995057
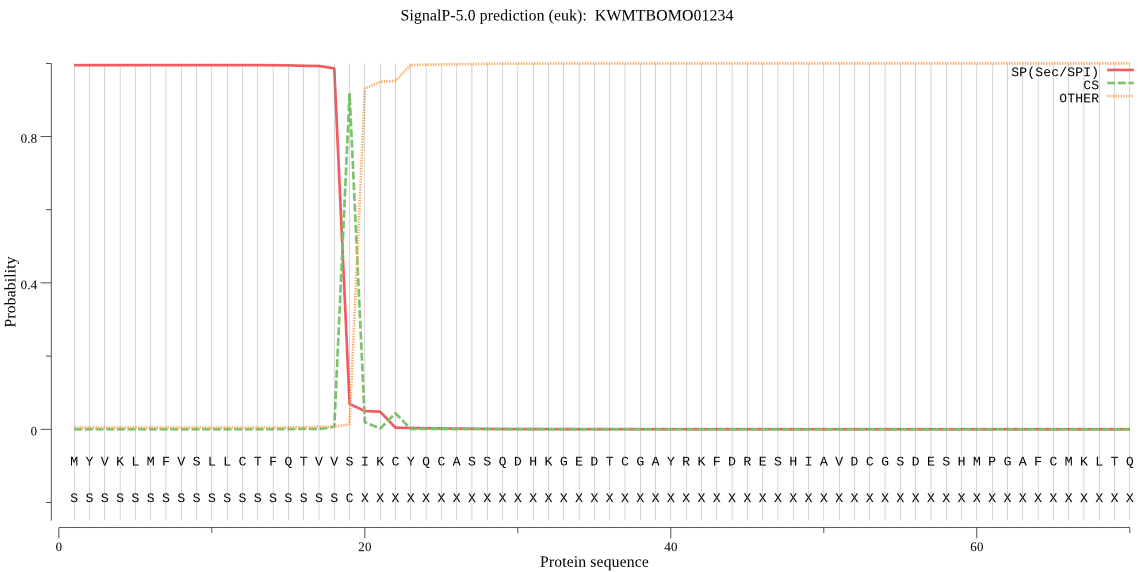
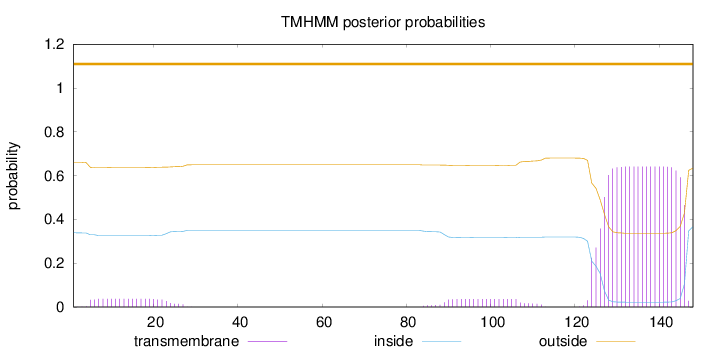
Length:
148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.8193
Exp number, first 60 AAs:
0.74635
Total prob of N-in:
0.33948
outside
1 - 148
Population Genetic Test Statistics
Pi
183.638976
Theta
183.862612
Tajima's D
-0.53259
CLR
0.681756
CSRT
0.23638818059097
Interpretation
Uncertain
