Gene
KWMTBOMO01233
Pre Gene Modal
BGIBMGA009034
Annotation
PREDICTED:_uncharacterized_protein_LOC105381223_[Plutella_xylostella]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
PlasmaMembrane Reliability : 2.897
Sequence
CDS
ATGGAAATTAACAAACTTAGTAATTTTGTTGGTTTTTTTATTATTTTTAGCTTAATGCCTCTCGCTCTAAGTATAAAATGTTGGAACTGCAGATCCAGCACCGATCCCAAATGCGCAGATCCTTTCGATAATAGTACAGTACCGATAACTGATTGCAATCAAGAAAAGGGACTTTCGCATTTGCCGGGAATGAAACCAAGCATGTGCCGCAAAATAAGACAAAAAGTTAATGGTGAATGGAGGTACTTCCGTGACTGTGCTTATTTGGGTGAAGTTGGCATTCAAGGAGATGAAAGGTTTTGTCTCATGAGGACTGGTACATACAATATATTTGTAGAATACTGTACTTGTAACAGTAAAGATGGTTGTAATTCTTCTTCAGTCATCAGTCCTTTACCTATATTCTTAGTTGTATTAGTTGCGTTAAGCTCTTTTAAGATAGGGCTCTAA
Protein
MEINKLSNFVGFFIIFSLMPLALSIKCWNCRSSTDPKCADPFDNSTVPITDCNQEKGLSHLPGMKPSMCRKIRQKVNGEWRYFRDCAYLGEVGIQGDERFCLMRTGTYNIFVEYCTCNSKDGCNSSSVISPLPIFLVVLVALSSFKIGL
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
H9JHN6
A0A2H1WTS3
A0A1E1W9B6
A0A2W1BSW2
A0A0N0PDK8
A0A194QID9
+ More
S4PL98 A0A224XZC1 A0A0N7Z879 T1I368 A0A026WLW1 K7J1R7 A0A232FCC3 A0A195CJU3 A0A0M9A643 A0A154PCQ5 A0A2R7W230 E2BK80 F4W418 A0A1Y0F467 A0A151X1G6 A0A088AWB2 A0A195B398 A0A158NK11 A0A2K8JRV4 A0A0L7QT56 A0A195DVV2 A0A2A3EF42 A0A146M1P2 A0A195EUH3 W5JGJ4 A0A182G4H1 A0A1B6FS97 A0A023EGZ6 B4MVF2 A0A2M4C284 A0A1B6JX96 A0A2M4AZ43 B0X7N1 A0A2M4AZ32 A0A2M4AZ13 A0A2J7QW69 A0A1I8N1F4 A0A1B6LI67 A0A084W7P3 A0A034WML7 A0A0K8UC64 Q9VKA0 A0A2M3ZBW4 A0A1I8QEK6 A0A2P8ZEX6 U5ES17 A0A3B0JA99 A0A182IIX4 A0A0A1XG12 A0A182RK48 A0A1Q3F053 A0A1Q3EZY7 B3N3V9 A0A1A9Y496 A0A1B0B2B1 B4KEG7 B4Q3R7 A0A182NGP8 B4P200 A0A1B0AAM6 A0A182QNP4 A0A1A9VA75 A0A1W4VEZ0 Q29M72 B4G9N0 A0A1B0FHK6 Q17CL2 A0A182MIE0 B3MNV3 A0A1B0C8R2 W8C620 A0A182F362 A0A1L8DNU2 B4LSX1 A0A0M4EAH2 A0A0L0BT32 A0A1L8DXX8 A0A1Y9IVK8 A0A182S9E9 A0A2C9GS70 A0A182UAP7 Q7Q8G6 B4JCB7 A0A182UYD7 A0A1A9WZK9 A0A182PVG4 A0A182X5W0 A0A182JZ17 A0A1W4XEL2 A0A336K8A4 D6WXR9
S4PL98 A0A224XZC1 A0A0N7Z879 T1I368 A0A026WLW1 K7J1R7 A0A232FCC3 A0A195CJU3 A0A0M9A643 A0A154PCQ5 A0A2R7W230 E2BK80 F4W418 A0A1Y0F467 A0A151X1G6 A0A088AWB2 A0A195B398 A0A158NK11 A0A2K8JRV4 A0A0L7QT56 A0A195DVV2 A0A2A3EF42 A0A146M1P2 A0A195EUH3 W5JGJ4 A0A182G4H1 A0A1B6FS97 A0A023EGZ6 B4MVF2 A0A2M4C284 A0A1B6JX96 A0A2M4AZ43 B0X7N1 A0A2M4AZ32 A0A2M4AZ13 A0A2J7QW69 A0A1I8N1F4 A0A1B6LI67 A0A084W7P3 A0A034WML7 A0A0K8UC64 Q9VKA0 A0A2M3ZBW4 A0A1I8QEK6 A0A2P8ZEX6 U5ES17 A0A3B0JA99 A0A182IIX4 A0A0A1XG12 A0A182RK48 A0A1Q3F053 A0A1Q3EZY7 B3N3V9 A0A1A9Y496 A0A1B0B2B1 B4KEG7 B4Q3R7 A0A182NGP8 B4P200 A0A1B0AAM6 A0A182QNP4 A0A1A9VA75 A0A1W4VEZ0 Q29M72 B4G9N0 A0A1B0FHK6 Q17CL2 A0A182MIE0 B3MNV3 A0A1B0C8R2 W8C620 A0A182F362 A0A1L8DNU2 B4LSX1 A0A0M4EAH2 A0A0L0BT32 A0A1L8DXX8 A0A1Y9IVK8 A0A182S9E9 A0A2C9GS70 A0A182UAP7 Q7Q8G6 B4JCB7 A0A182UYD7 A0A1A9WZK9 A0A182PVG4 A0A182X5W0 A0A182JZ17 A0A1W4XEL2 A0A336K8A4 D6WXR9
Pubmed
19121390
28756777
26354079
23622113
27129103
24508170
+ More
30249741 20075255 28648823 20798317 21719571 21347285 26823975 20920257 23761445 26483478 24945155 17994087 25315136 24438588 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25830018 22936249 17550304 15632085 17510324 24495485 26108605 12364791 14747013 17210077 18362917 19820115
30249741 20075255 28648823 20798317 21719571 21347285 26823975 20920257 23761445 26483478 24945155 17994087 25315136 24438588 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25830018 22936249 17550304 15632085 17510324 24495485 26108605 12364791 14747013 17210077 18362917 19820115
EMBL
BABH01012984
ODYU01010604
SOQ55824.1
GDQN01007603
JAT83451.1
KZ149910
+ More
PZC78148.1 KQ460317 KPJ16012.1 KQ458756 KPJ05282.1 GAIX01004140 JAA88420.1 GFTR01002456 JAW13970.1 GDKW01004099 JAI52496.1 ACPB03014814 KK107153 QOIP01000012 EZA56953.1 RLU15790.1 NNAY01000427 OXU28481.1 KQ977642 KYN01000.1 KQ435727 KOX78005.1 KQ434870 KZC09603.1 KK854233 PTY13488.1 GL448770 EFN83877.1 GL887491 EGI71014.1 KX950845 ARU12055.1 KQ982585 KYQ54249.1 KQ976625 KYM78968.1 ADTU01018617 KY031019 ATU82770.1 KQ414755 KOC61805.1 KQ980295 KYN16872.1 KZ288263 PBC30340.1 GDHC01005853 JAQ12776.1 KQ981979 KYN31529.1 ADMH02001597 ETN61914.1 JXUM01042353 KQ561321 KXJ78986.1 GECZ01025600 GECZ01016709 JAS44169.1 JAS53060.1 GAPW01005499 JAC08099.1 CH963857 EDW76497.1 GGFJ01010305 MBW59446.1 GECU01004193 JAT03514.1 GGFK01012752 MBW46073.1 DS232456 EDS42043.1 GGFK01012709 MBW46030.1 GGFK01012710 MBW46031.1 NEVH01009767 PNF32830.1 GEBQ01016628 JAT23349.1 ATLV01021269 KE525315 KFB46237.1 GAKP01003370 JAC55582.1 GDHF01028364 JAI23950.1 AE014134 BT044115 KX531517 AAF53178.1 ACH92180.1 ANY27327.1 GGFM01005209 MBW25960.1 PYGN01000077 PSN55054.1 GANO01003458 JAB56413.1 OUUW01000004 SPP79217.1 GBXI01004406 JAD09886.1 GFDL01014113 JAV20932.1 GFDL01014170 JAV20875.1 CH954177 EDV58811.1 JXJN01007544 CH933807 EDW12935.1 CM000361 CM002910 EDX04792.1 KMY89895.1 CM000157 EDW88171.1 AXCN02002080 CH379060 EAL33822.1 CH479180 EDW29060.1 CCAG010008329 CH477307 EAT44085.1 AXCM01014743 CH902620 EDV32140.1 AJWK01001280 GAMC01008761 JAB97794.1 GFDF01005941 JAV08143.1 CH940649 EDW64882.1 CP012523 ALC39388.1 JRES01001399 KNC23181.1 GFDF01002781 JAV11303.1 APCN01000707 AAAB01008944 EAA10235.3 CH916368 EDW04150.1 UFQS01000114 UFQT01000114 SSW99848.1 SSX20228.1 KQ971362 EFA08898.1
PZC78148.1 KQ460317 KPJ16012.1 KQ458756 KPJ05282.1 GAIX01004140 JAA88420.1 GFTR01002456 JAW13970.1 GDKW01004099 JAI52496.1 ACPB03014814 KK107153 QOIP01000012 EZA56953.1 RLU15790.1 NNAY01000427 OXU28481.1 KQ977642 KYN01000.1 KQ435727 KOX78005.1 KQ434870 KZC09603.1 KK854233 PTY13488.1 GL448770 EFN83877.1 GL887491 EGI71014.1 KX950845 ARU12055.1 KQ982585 KYQ54249.1 KQ976625 KYM78968.1 ADTU01018617 KY031019 ATU82770.1 KQ414755 KOC61805.1 KQ980295 KYN16872.1 KZ288263 PBC30340.1 GDHC01005853 JAQ12776.1 KQ981979 KYN31529.1 ADMH02001597 ETN61914.1 JXUM01042353 KQ561321 KXJ78986.1 GECZ01025600 GECZ01016709 JAS44169.1 JAS53060.1 GAPW01005499 JAC08099.1 CH963857 EDW76497.1 GGFJ01010305 MBW59446.1 GECU01004193 JAT03514.1 GGFK01012752 MBW46073.1 DS232456 EDS42043.1 GGFK01012709 MBW46030.1 GGFK01012710 MBW46031.1 NEVH01009767 PNF32830.1 GEBQ01016628 JAT23349.1 ATLV01021269 KE525315 KFB46237.1 GAKP01003370 JAC55582.1 GDHF01028364 JAI23950.1 AE014134 BT044115 KX531517 AAF53178.1 ACH92180.1 ANY27327.1 GGFM01005209 MBW25960.1 PYGN01000077 PSN55054.1 GANO01003458 JAB56413.1 OUUW01000004 SPP79217.1 GBXI01004406 JAD09886.1 GFDL01014113 JAV20932.1 GFDL01014170 JAV20875.1 CH954177 EDV58811.1 JXJN01007544 CH933807 EDW12935.1 CM000361 CM002910 EDX04792.1 KMY89895.1 CM000157 EDW88171.1 AXCN02002080 CH379060 EAL33822.1 CH479180 EDW29060.1 CCAG010008329 CH477307 EAT44085.1 AXCM01014743 CH902620 EDV32140.1 AJWK01001280 GAMC01008761 JAB97794.1 GFDF01005941 JAV08143.1 CH940649 EDW64882.1 CP012523 ALC39388.1 JRES01001399 KNC23181.1 GFDF01002781 JAV11303.1 APCN01000707 AAAB01008944 EAA10235.3 CH916368 EDW04150.1 UFQS01000114 UFQT01000114 SSW99848.1 SSX20228.1 KQ971362 EFA08898.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000015103
UP000053097
UP000279307
+ More
UP000002358 UP000215335 UP000078542 UP000053105 UP000076502 UP000008237 UP000007755 UP000075809 UP000005203 UP000078540 UP000005205 UP000053825 UP000078492 UP000242457 UP000078541 UP000000673 UP000069940 UP000249989 UP000007798 UP000002320 UP000235965 UP000095301 UP000030765 UP000000803 UP000095300 UP000245037 UP000268350 UP000075880 UP000075900 UP000008711 UP000092443 UP000092460 UP000009192 UP000000304 UP000075884 UP000002282 UP000092445 UP000075886 UP000078200 UP000192221 UP000001819 UP000008744 UP000092444 UP000008820 UP000075883 UP000007801 UP000092461 UP000069272 UP000008792 UP000092553 UP000037069 UP000075920 UP000075901 UP000075840 UP000075902 UP000007062 UP000001070 UP000075903 UP000091820 UP000075885 UP000076407 UP000075881 UP000192223 UP000007266
UP000002358 UP000215335 UP000078542 UP000053105 UP000076502 UP000008237 UP000007755 UP000075809 UP000005203 UP000078540 UP000005205 UP000053825 UP000078492 UP000242457 UP000078541 UP000000673 UP000069940 UP000249989 UP000007798 UP000002320 UP000235965 UP000095301 UP000030765 UP000000803 UP000095300 UP000245037 UP000268350 UP000075880 UP000075900 UP000008711 UP000092443 UP000092460 UP000009192 UP000000304 UP000075884 UP000002282 UP000092445 UP000075886 UP000078200 UP000192221 UP000001819 UP000008744 UP000092444 UP000008820 UP000075883 UP000007801 UP000092461 UP000069272 UP000008792 UP000092553 UP000037069 UP000075920 UP000075901 UP000075840 UP000075902 UP000007062 UP000001070 UP000075903 UP000091820 UP000075885 UP000076407 UP000075881 UP000192223 UP000007266
Pfam
PF17064 QVR
Interpro
IPR031424
QVR
ProteinModelPortal
H9JHN6
A0A2H1WTS3
A0A1E1W9B6
A0A2W1BSW2
A0A0N0PDK8
A0A194QID9
+ More
S4PL98 A0A224XZC1 A0A0N7Z879 T1I368 A0A026WLW1 K7J1R7 A0A232FCC3 A0A195CJU3 A0A0M9A643 A0A154PCQ5 A0A2R7W230 E2BK80 F4W418 A0A1Y0F467 A0A151X1G6 A0A088AWB2 A0A195B398 A0A158NK11 A0A2K8JRV4 A0A0L7QT56 A0A195DVV2 A0A2A3EF42 A0A146M1P2 A0A195EUH3 W5JGJ4 A0A182G4H1 A0A1B6FS97 A0A023EGZ6 B4MVF2 A0A2M4C284 A0A1B6JX96 A0A2M4AZ43 B0X7N1 A0A2M4AZ32 A0A2M4AZ13 A0A2J7QW69 A0A1I8N1F4 A0A1B6LI67 A0A084W7P3 A0A034WML7 A0A0K8UC64 Q9VKA0 A0A2M3ZBW4 A0A1I8QEK6 A0A2P8ZEX6 U5ES17 A0A3B0JA99 A0A182IIX4 A0A0A1XG12 A0A182RK48 A0A1Q3F053 A0A1Q3EZY7 B3N3V9 A0A1A9Y496 A0A1B0B2B1 B4KEG7 B4Q3R7 A0A182NGP8 B4P200 A0A1B0AAM6 A0A182QNP4 A0A1A9VA75 A0A1W4VEZ0 Q29M72 B4G9N0 A0A1B0FHK6 Q17CL2 A0A182MIE0 B3MNV3 A0A1B0C8R2 W8C620 A0A182F362 A0A1L8DNU2 B4LSX1 A0A0M4EAH2 A0A0L0BT32 A0A1L8DXX8 A0A1Y9IVK8 A0A182S9E9 A0A2C9GS70 A0A182UAP7 Q7Q8G6 B4JCB7 A0A182UYD7 A0A1A9WZK9 A0A182PVG4 A0A182X5W0 A0A182JZ17 A0A1W4XEL2 A0A336K8A4 D6WXR9
S4PL98 A0A224XZC1 A0A0N7Z879 T1I368 A0A026WLW1 K7J1R7 A0A232FCC3 A0A195CJU3 A0A0M9A643 A0A154PCQ5 A0A2R7W230 E2BK80 F4W418 A0A1Y0F467 A0A151X1G6 A0A088AWB2 A0A195B398 A0A158NK11 A0A2K8JRV4 A0A0L7QT56 A0A195DVV2 A0A2A3EF42 A0A146M1P2 A0A195EUH3 W5JGJ4 A0A182G4H1 A0A1B6FS97 A0A023EGZ6 B4MVF2 A0A2M4C284 A0A1B6JX96 A0A2M4AZ43 B0X7N1 A0A2M4AZ32 A0A2M4AZ13 A0A2J7QW69 A0A1I8N1F4 A0A1B6LI67 A0A084W7P3 A0A034WML7 A0A0K8UC64 Q9VKA0 A0A2M3ZBW4 A0A1I8QEK6 A0A2P8ZEX6 U5ES17 A0A3B0JA99 A0A182IIX4 A0A0A1XG12 A0A182RK48 A0A1Q3F053 A0A1Q3EZY7 B3N3V9 A0A1A9Y496 A0A1B0B2B1 B4KEG7 B4Q3R7 A0A182NGP8 B4P200 A0A1B0AAM6 A0A182QNP4 A0A1A9VA75 A0A1W4VEZ0 Q29M72 B4G9N0 A0A1B0FHK6 Q17CL2 A0A182MIE0 B3MNV3 A0A1B0C8R2 W8C620 A0A182F362 A0A1L8DNU2 B4LSX1 A0A0M4EAH2 A0A0L0BT32 A0A1L8DXX8 A0A1Y9IVK8 A0A182S9E9 A0A2C9GS70 A0A182UAP7 Q7Q8G6 B4JCB7 A0A182UYD7 A0A1A9WZK9 A0A182PVG4 A0A182X5W0 A0A182JZ17 A0A1W4XEL2 A0A336K8A4 D6WXR9
Ontologies
GO
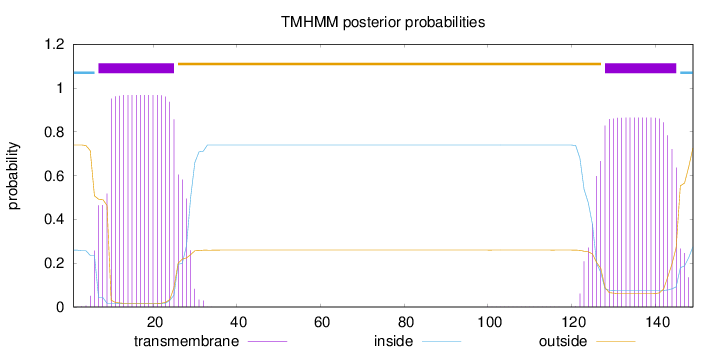
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 24,
Likelihood: 0.977823
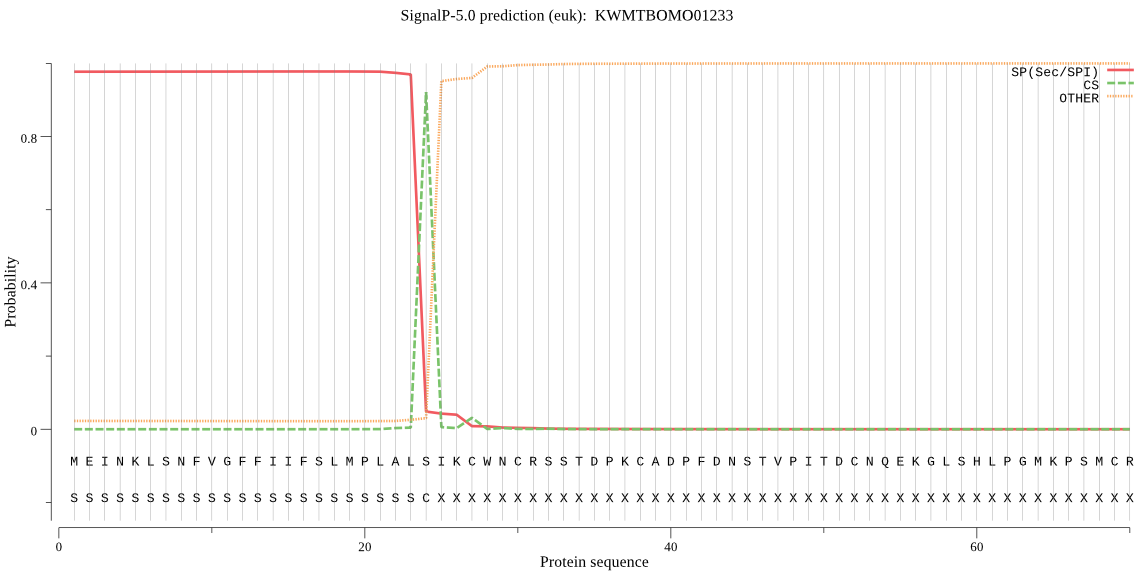
Length:
149
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
37.03738
Exp number, first 60 AAs:
19.15827
Total prob of N-in:
0.25984
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 25
outside
26 - 127
TMhelix
128 - 145
inside
146 - 149
Population Genetic Test Statistics
Pi
208.355379
Theta
183.515225
Tajima's D
0.495307
CLR
0.396024
CSRT
0.518174091295435
Interpretation
Uncertain
