Gene
KWMTBOMO01232 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008848
Annotation
putative_ly-6/neurotoxin_superfamily_member_1_[Operophtera_brumata]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 4.448
Sequence
CDS
ATGGCAAAACTATCGTTTCTTGTGCCCGTTTTGCTTCTTGCGCTATTCGTGAAGGACAGTGAACCAGTGAATTGTTGGGCGTGTTCGTCTAACGTGAATCCGCTGTGCAACGATCCTTTTAATATCCGTATCGACACCGGGAATTCTTATTTGTTCCGCCTCGAAAACTGCGACAAGAATGCTGGAGCTACCTTTCCTTATTTGACAGCTTCGAAGTCTGTATGCAAGAAAGAAAAGAAATACATTGATGGTGAATTAGTTGTGAGTCGTGGTTGCACCTGGAAACGCCAGGATGACTTCGAAGTCGGTTGCCCTACTTCCAGAAACGAAGCAAACGAAGTGAATCTCTTCTGTCAGACCTGTGACTACGACGGATGCAACGGAGCCGCTACCATCGGCAGGACGATCGCTCTATTGCTCGCACCTCTCAGCCTCCTCTTGTTGAAGTAA
Protein
MAKLSFLVPVLLLALFVKDSEPVNCWACSSNVNPLCNDPFNIRIDTGNSYLFRLENCDKNAGATFPYLTASKSVCKKEKKYIDGELVVSRGCTWKRQDDFEVGCPTSRNEANEVNLFCQTCDYDGCNGAATIGRTIALLLAPLSLLLLK
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
H9JH51
A0A2W1BY38
A0A2H1X2D0
A0A0L7L7W8
I4DJK1
A0A194QIQ9
+ More
A0A212F7X5 S4P8V1 A0A1E1W9R6 A0A069DPK0 A0A224XN13 A0A0P4VKI5 A0A0V0G953 Q1HQZ0 A0A2S2NT63 U5EDB7 A0A023EFU8 A0A182G4H2 C4WW54 B0X7N2 A0A2S2Q6L0 A0A0A9X2S6 A0A1Q3FZI3 A0A1Q3FZJ7 A0A146MCS9 A0A1B0CML5 A0A336K8A4 A0A1L8DNZ8 A0A1J1IL94 T1D554 A0A084W7P2 A0A1B6DS46 B3NKY0 R4UNF8 C5IWP3 B4IFH2 B4Q3V6 Q9VII5 B4P724 A0A0K8VK42 A0A034VLL4 A0A1I8MED2 A0A067R4K6 W8B283 A0A182U3Y2 A0A2M4A5I1 A0A2M4A553 A0A1S4GYT8 A0A2C9GS45 A0A182UND5 A0A158NNF8 Q7Q8G7 A0A2M4DN25 A0A2M4A497 A0A0L0CG35 A0A3L8DS81 E1ZV82 A0A151WUG8 A0A182QTB6 A0A2M3YXU6 A0A2M3ZE83 A0A0A1WPV7 A0A1W4XP76 A0A2M3YWC5 A0A0C5AYN4 A0A182F362 A0A336KJT5 A0A3B0K1X1 T1E9L8 A0A1Y9IWC7 A0A2M4C2D0 A0A2M4C2A7 B5DIR0 B4G725 A0A2M4C2B6 A0A2M4C1V1 V9ILT4 A0A088AHD2 A0A195CCW9 A0A1W4UW05 E9IIU0 E0VVY9 A0A182RK49 B4MV59 A0A2A3E573 A0A0M4E781 A0A0L7R443 A0A1I8PHK9 B3MU82 A0A182PVG4 A0A0M8ZZ44 B4LUN4 B4JD94 A0A182JZ17 B4KL10 A0A0Q9XII0 A0A1B0GAJ1 A0A154PAQ1
A0A212F7X5 S4P8V1 A0A1E1W9R6 A0A069DPK0 A0A224XN13 A0A0P4VKI5 A0A0V0G953 Q1HQZ0 A0A2S2NT63 U5EDB7 A0A023EFU8 A0A182G4H2 C4WW54 B0X7N2 A0A2S2Q6L0 A0A0A9X2S6 A0A1Q3FZI3 A0A1Q3FZJ7 A0A146MCS9 A0A1B0CML5 A0A336K8A4 A0A1L8DNZ8 A0A1J1IL94 T1D554 A0A084W7P2 A0A1B6DS46 B3NKY0 R4UNF8 C5IWP3 B4IFH2 B4Q3V6 Q9VII5 B4P724 A0A0K8VK42 A0A034VLL4 A0A1I8MED2 A0A067R4K6 W8B283 A0A182U3Y2 A0A2M4A5I1 A0A2M4A553 A0A1S4GYT8 A0A2C9GS45 A0A182UND5 A0A158NNF8 Q7Q8G7 A0A2M4DN25 A0A2M4A497 A0A0L0CG35 A0A3L8DS81 E1ZV82 A0A151WUG8 A0A182QTB6 A0A2M3YXU6 A0A2M3ZE83 A0A0A1WPV7 A0A1W4XP76 A0A2M3YWC5 A0A0C5AYN4 A0A182F362 A0A336KJT5 A0A3B0K1X1 T1E9L8 A0A1Y9IWC7 A0A2M4C2D0 A0A2M4C2A7 B5DIR0 B4G725 A0A2M4C2B6 A0A2M4C1V1 V9ILT4 A0A088AHD2 A0A195CCW9 A0A1W4UW05 E9IIU0 E0VVY9 A0A182RK49 B4MV59 A0A2A3E573 A0A0M4E781 A0A0L7R443 A0A1I8PHK9 B3MU82 A0A182PVG4 A0A0M8ZZ44 B4LUN4 B4JD94 A0A182JZ17 B4KL10 A0A0Q9XII0 A0A1B0GAJ1 A0A154PAQ1
Pubmed
19121390
28756777
26227816
22651552
26354079
22118469
+ More
23622113 26334808 27129103 17204158 17510324 24945155 26483478 25401762 26823975 24330624 24438588 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 25315136 24845553 24495485 12364791 21347285 26108605 30249741 20798317 25830018 15632085 21282665 20566863 18057021
23622113 26334808 27129103 17204158 17510324 24945155 26483478 25401762 26823975 24330624 24438588 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 25315136 24845553 24495485 12364791 21347285 26108605 30249741 20798317 25830018 15632085 21282665 20566863 18057021
EMBL
BABH01012984
KZ149910
PZC78147.1
ODYU01012936
SOQ59481.1
JTDY01002449
+ More
KOB71391.1 AK401469 BAM18091.1 KQ458756 KPJ05284.1 AGBW02009823 OWR49844.1 GAIX01007117 JAA85443.1 GDQN01007304 JAT83750.1 GBGD01003342 JAC85547.1 GFTR01002540 JAW13886.1 GDKW01003584 JAI53011.1 GECL01001598 JAP04526.1 DQ440304 CH477307 ABF18337.1 EAT44084.1 GGMR01007726 MBY20345.1 GANO01004834 JAB55037.1 GAPW01005310 JAC08288.1 JXUM01042357 JXUM01042358 JXUM01042359 JXUM01042360 KQ561321 KXJ78987.1 ABLF02036169 AK341857 BAH72124.1 DS232456 EDS42044.1 GGMS01004038 MBY73241.1 GBHO01029638 GBHO01029637 GBHO01029636 GBHO01029635 GBHO01029634 GBHO01029633 GBRD01013774 GBRD01013773 GBRD01013772 GBRD01013770 JAG13966.1 JAG13967.1 JAG13968.1 JAG13969.1 JAG13970.1 JAG13971.1 JAG52052.1 GFDL01002092 JAV32953.1 GFDL01002067 JAV32978.1 GDHC01002289 JAQ16340.1 AJWK01018884 AJWK01018885 AJWK01018886 AJWK01018887 UFQS01000114 UFQT01000114 SSW99848.1 SSX20228.1 GFDF01005886 JAV08198.1 CVRI01000055 CRL01013.1 GALA01000678 JAA94174.1 ATLV01021269 KE525315 KFB46236.1 GEDC01008796 JAS28502.1 CH954179 EDV54503.1 KC740871 AGM32695.1 FJ968776 ACR56003.1 CH480833 EDW46394.1 CM000361 CM002910 EDX05672.1 KMY91231.1 AE014134 AY075543 KX531351 AAF53934.3 AAL68350.1 AHN54627.1 ANY27161.1 CM000158 EDW89993.1 GDHF01033478 GDHF01013082 JAI18836.1 JAI39232.1 GAKP01014771 JAC44181.1 KK852699 KDR18171.1 GAMC01019179 JAB87376.1 GGFK01002669 MBW35990.1 GGFK01002616 MBW35937.1 AAAB01008944 APCN01000707 ADTU01021428 ADTU01021429 ADTU01021430 ADTU01021431 ADTU01021432 ADTU01021433 ADTU01021434 ADTU01021435 ADTU01021436 ADTU01021437 EAA10065.4 GGFL01014792 MBW78970.1 GGFK01002313 MBW35634.1 JRES01000438 KNC31210.1 QOIP01000004 RLU23290.1 GL434441 EFN74941.1 KQ982730 KYQ51486.1 AXCN02002080 GGFM01000336 MBW21087.1 GGFM01006095 MBW26846.1 GBXI01013596 JAD00696.1 GGFF01000090 MBW20557.1 KM974262 AJL35192.1 UFQS01000518 UFQT01000518 SSX04541.1 SSX24904.1 OUUW01000004 SPP79596.1 GAMD01001227 JAB00364.1 GGFJ01010302 MBW59443.1 GGFJ01010301 MBW59442.1 CH379060 EDY70202.1 CH479180 EDW29224.1 GGFJ01010170 MBW59311.1 GGFJ01010169 MBW59310.1 JR050453 AEY61284.1 KQ977935 KYM98565.1 GL763529 EFZ19513.1 DS235816 EEB17545.1 CH963857 EDW76404.1 KZ288365 PBC26840.1 CP012523 ALC39566.1 ALC40334.1 KQ414659 KOC65618.1 CH902624 EDV33411.1 KPU74465.1 KQ435819 KOX72519.1 CH940649 EDW64220.1 CH916368 EDW03267.1 CH933807 EDW12760.2 KRG03411.1 CCAG010013001 CCAG010013002 CCAG010013003 KQ434860 KZC08922.1
KOB71391.1 AK401469 BAM18091.1 KQ458756 KPJ05284.1 AGBW02009823 OWR49844.1 GAIX01007117 JAA85443.1 GDQN01007304 JAT83750.1 GBGD01003342 JAC85547.1 GFTR01002540 JAW13886.1 GDKW01003584 JAI53011.1 GECL01001598 JAP04526.1 DQ440304 CH477307 ABF18337.1 EAT44084.1 GGMR01007726 MBY20345.1 GANO01004834 JAB55037.1 GAPW01005310 JAC08288.1 JXUM01042357 JXUM01042358 JXUM01042359 JXUM01042360 KQ561321 KXJ78987.1 ABLF02036169 AK341857 BAH72124.1 DS232456 EDS42044.1 GGMS01004038 MBY73241.1 GBHO01029638 GBHO01029637 GBHO01029636 GBHO01029635 GBHO01029634 GBHO01029633 GBRD01013774 GBRD01013773 GBRD01013772 GBRD01013770 JAG13966.1 JAG13967.1 JAG13968.1 JAG13969.1 JAG13970.1 JAG13971.1 JAG52052.1 GFDL01002092 JAV32953.1 GFDL01002067 JAV32978.1 GDHC01002289 JAQ16340.1 AJWK01018884 AJWK01018885 AJWK01018886 AJWK01018887 UFQS01000114 UFQT01000114 SSW99848.1 SSX20228.1 GFDF01005886 JAV08198.1 CVRI01000055 CRL01013.1 GALA01000678 JAA94174.1 ATLV01021269 KE525315 KFB46236.1 GEDC01008796 JAS28502.1 CH954179 EDV54503.1 KC740871 AGM32695.1 FJ968776 ACR56003.1 CH480833 EDW46394.1 CM000361 CM002910 EDX05672.1 KMY91231.1 AE014134 AY075543 KX531351 AAF53934.3 AAL68350.1 AHN54627.1 ANY27161.1 CM000158 EDW89993.1 GDHF01033478 GDHF01013082 JAI18836.1 JAI39232.1 GAKP01014771 JAC44181.1 KK852699 KDR18171.1 GAMC01019179 JAB87376.1 GGFK01002669 MBW35990.1 GGFK01002616 MBW35937.1 AAAB01008944 APCN01000707 ADTU01021428 ADTU01021429 ADTU01021430 ADTU01021431 ADTU01021432 ADTU01021433 ADTU01021434 ADTU01021435 ADTU01021436 ADTU01021437 EAA10065.4 GGFL01014792 MBW78970.1 GGFK01002313 MBW35634.1 JRES01000438 KNC31210.1 QOIP01000004 RLU23290.1 GL434441 EFN74941.1 KQ982730 KYQ51486.1 AXCN02002080 GGFM01000336 MBW21087.1 GGFM01006095 MBW26846.1 GBXI01013596 JAD00696.1 GGFF01000090 MBW20557.1 KM974262 AJL35192.1 UFQS01000518 UFQT01000518 SSX04541.1 SSX24904.1 OUUW01000004 SPP79596.1 GAMD01001227 JAB00364.1 GGFJ01010302 MBW59443.1 GGFJ01010301 MBW59442.1 CH379060 EDY70202.1 CH479180 EDW29224.1 GGFJ01010170 MBW59311.1 GGFJ01010169 MBW59310.1 JR050453 AEY61284.1 KQ977935 KYM98565.1 GL763529 EFZ19513.1 DS235816 EEB17545.1 CH963857 EDW76404.1 KZ288365 PBC26840.1 CP012523 ALC39566.1 ALC40334.1 KQ414659 KOC65618.1 CH902624 EDV33411.1 KPU74465.1 KQ435819 KOX72519.1 CH940649 EDW64220.1 CH916368 EDW03267.1 CH933807 EDW12760.2 KRG03411.1 CCAG010013001 CCAG010013002 CCAG010013003 KQ434860 KZC08922.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000007151
UP000008820
UP000069940
+ More
UP000249989 UP000007819 UP000002320 UP000092461 UP000183832 UP000030765 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000095301 UP000027135 UP000075902 UP000075840 UP000075903 UP000005205 UP000007062 UP000037069 UP000279307 UP000000311 UP000075809 UP000075886 UP000192223 UP000069272 UP000268350 UP000075920 UP000001819 UP000008744 UP000005203 UP000078542 UP000192221 UP000009046 UP000075900 UP000007798 UP000242457 UP000092553 UP000053825 UP000095300 UP000007801 UP000075885 UP000053105 UP000008792 UP000001070 UP000075881 UP000009192 UP000092444 UP000076502
UP000249989 UP000007819 UP000002320 UP000092461 UP000183832 UP000030765 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000095301 UP000027135 UP000075902 UP000075840 UP000075903 UP000005205 UP000007062 UP000037069 UP000279307 UP000000311 UP000075809 UP000075886 UP000192223 UP000069272 UP000268350 UP000075920 UP000001819 UP000008744 UP000005203 UP000078542 UP000192221 UP000009046 UP000075900 UP000007798 UP000242457 UP000092553 UP000053825 UP000095300 UP000007801 UP000075885 UP000053105 UP000008792 UP000001070 UP000075881 UP000009192 UP000092444 UP000076502
Pfam
PF17064 QVR
Interpro
IPR031424
QVR
ProteinModelPortal
H9JH51
A0A2W1BY38
A0A2H1X2D0
A0A0L7L7W8
I4DJK1
A0A194QIQ9
+ More
A0A212F7X5 S4P8V1 A0A1E1W9R6 A0A069DPK0 A0A224XN13 A0A0P4VKI5 A0A0V0G953 Q1HQZ0 A0A2S2NT63 U5EDB7 A0A023EFU8 A0A182G4H2 C4WW54 B0X7N2 A0A2S2Q6L0 A0A0A9X2S6 A0A1Q3FZI3 A0A1Q3FZJ7 A0A146MCS9 A0A1B0CML5 A0A336K8A4 A0A1L8DNZ8 A0A1J1IL94 T1D554 A0A084W7P2 A0A1B6DS46 B3NKY0 R4UNF8 C5IWP3 B4IFH2 B4Q3V6 Q9VII5 B4P724 A0A0K8VK42 A0A034VLL4 A0A1I8MED2 A0A067R4K6 W8B283 A0A182U3Y2 A0A2M4A5I1 A0A2M4A553 A0A1S4GYT8 A0A2C9GS45 A0A182UND5 A0A158NNF8 Q7Q8G7 A0A2M4DN25 A0A2M4A497 A0A0L0CG35 A0A3L8DS81 E1ZV82 A0A151WUG8 A0A182QTB6 A0A2M3YXU6 A0A2M3ZE83 A0A0A1WPV7 A0A1W4XP76 A0A2M3YWC5 A0A0C5AYN4 A0A182F362 A0A336KJT5 A0A3B0K1X1 T1E9L8 A0A1Y9IWC7 A0A2M4C2D0 A0A2M4C2A7 B5DIR0 B4G725 A0A2M4C2B6 A0A2M4C1V1 V9ILT4 A0A088AHD2 A0A195CCW9 A0A1W4UW05 E9IIU0 E0VVY9 A0A182RK49 B4MV59 A0A2A3E573 A0A0M4E781 A0A0L7R443 A0A1I8PHK9 B3MU82 A0A182PVG4 A0A0M8ZZ44 B4LUN4 B4JD94 A0A182JZ17 B4KL10 A0A0Q9XII0 A0A1B0GAJ1 A0A154PAQ1
A0A212F7X5 S4P8V1 A0A1E1W9R6 A0A069DPK0 A0A224XN13 A0A0P4VKI5 A0A0V0G953 Q1HQZ0 A0A2S2NT63 U5EDB7 A0A023EFU8 A0A182G4H2 C4WW54 B0X7N2 A0A2S2Q6L0 A0A0A9X2S6 A0A1Q3FZI3 A0A1Q3FZJ7 A0A146MCS9 A0A1B0CML5 A0A336K8A4 A0A1L8DNZ8 A0A1J1IL94 T1D554 A0A084W7P2 A0A1B6DS46 B3NKY0 R4UNF8 C5IWP3 B4IFH2 B4Q3V6 Q9VII5 B4P724 A0A0K8VK42 A0A034VLL4 A0A1I8MED2 A0A067R4K6 W8B283 A0A182U3Y2 A0A2M4A5I1 A0A2M4A553 A0A1S4GYT8 A0A2C9GS45 A0A182UND5 A0A158NNF8 Q7Q8G7 A0A2M4DN25 A0A2M4A497 A0A0L0CG35 A0A3L8DS81 E1ZV82 A0A151WUG8 A0A182QTB6 A0A2M3YXU6 A0A2M3ZE83 A0A0A1WPV7 A0A1W4XP76 A0A2M3YWC5 A0A0C5AYN4 A0A182F362 A0A336KJT5 A0A3B0K1X1 T1E9L8 A0A1Y9IWC7 A0A2M4C2D0 A0A2M4C2A7 B5DIR0 B4G725 A0A2M4C2B6 A0A2M4C1V1 V9ILT4 A0A088AHD2 A0A195CCW9 A0A1W4UW05 E9IIU0 E0VVY9 A0A182RK49 B4MV59 A0A2A3E573 A0A0M4E781 A0A0L7R443 A0A1I8PHK9 B3MU82 A0A182PVG4 A0A0M8ZZ44 B4LUN4 B4JD94 A0A182JZ17 B4KL10 A0A0Q9XII0 A0A1B0GAJ1 A0A154PAQ1
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 22,
Likelihood: 0.998219
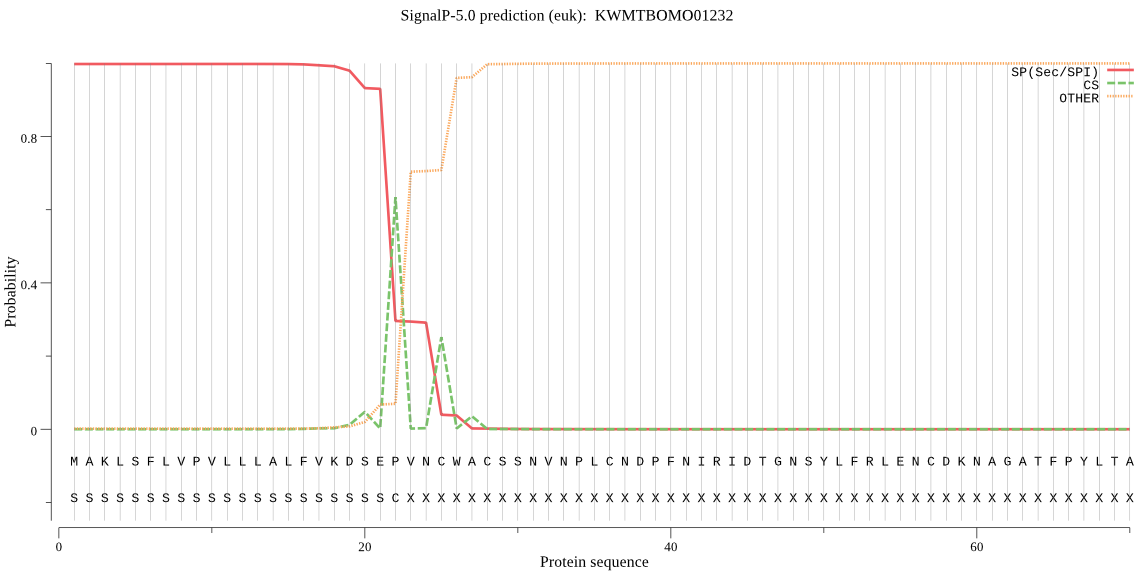
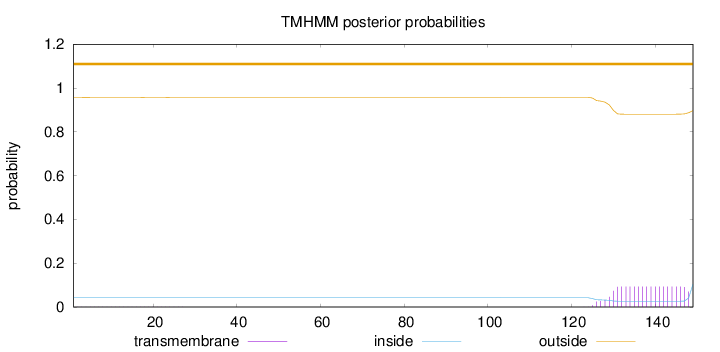
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.88286
Exp number, first 60 AAs:
0.01573
Total prob of N-in:
0.04256
outside
1 - 149
Population Genetic Test Statistics
Pi
219.772865
Theta
171.207314
Tajima's D
0.54875
CLR
0.432339
CSRT
0.527273636318184
Interpretation
Uncertain
