Gene
KWMTBOMO01229 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008847
Annotation
PREDICTED:_uncharacterized_protein_LOC101743420_[Bombyx_mori]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 4.474
Sequence
CDS
ATGGCAAAAAGTGTAGCATTCGCCTTCGCTGTTCTACTAGCGCTATTCGAAACAGGCTATTGCATCAAGTGCTACCAATGTAACAGTGAACAGGATAAGAACTGCGGGGACCCGTTCAAGAGCGCCAAACCCCCAGTAGAATGCAACACCCAGGATTCGATTAACTTCAATACGCTCTACCTGCGCAACATTCTGCCTGTCGAAGTCTTGAACAGCGTTACGGGAGCTCCTAGATACTGTCACAAGATCGTCATGAAGAGCGGAACTGTTGTCCGTACTTGCTTGGATGTCAACCCCAACGATTCCCAGCACACCTGCCGAGTTGTCGAGCTCGCCTCCAACACCGCGATCGCTGACAGCGCGAAGGTCAAGTCCTGCGCTGTGTGCAACAAGGACAACTGTAACGGAGCTGGATCTATTTCCTTCTCTCTCCCACTGGCCACATTCGCTCTGATCGCTACGTATTTCGTTTTCAAACAATAA
Protein
MAKSVAFAFAVLLALFETGYCIKCYQCNSEQDKNCGDPFKSAKPPVECNTQDSINFNTLYLRNILPVEVLNSVTGAPRYCHKIVMKSGTVVRTCLDVNPNDSQHTCRVVELASNTAIADSAKVKSCAVCNKDNCNGAGSISFSLPLATFALIATYFVFKQ
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
H9JH50
A0A2A4J434
A0A2W1BWX5
I4DL69
I4DMY3
A0A194QK48
+ More
A0A0N1IH29 A0A1E1W402 S4P3Z0 A0A0V0G953 R4G540 A0A069DPK0 A0A224XN13 A0A0J9R4N1 A0A1I8M3Z3 B4MV65 B4IFH8 B4Q3W2 B5DIQ8 B4G719 A0A224XUK9 B4P719 A0A0V0G2Y8 A0A286P0W6 B4LUN1 T1IFT4 C5IWP3 A0A0P4VKI5 Q9VIH9 A0A1W4UVF0 A0A151JBA9 A0A023FD89 B4JD91 R4G7Y0 B3PAG6 A0A3B0K3I3 B3NKX1 A0A2P8YVI6 T1I2D3 E2J782 A0A1W4WPE1 E0VVY9 A0A0M4EEB1 A0A195EWX4 A0A067R4V8 B3MU77 R4UWG9 A0A067R6Z0 A0A182K0L5 A0A0L7R443 B3NKW9 A0A2J7QW69 A0A1S3D977 S4PL98 Q0MTF2 A0A195C5M5 B3NKX5 A0A182PPI2 B3PAG8 A0A182L1H1 A0NF51 A0A182HUP4 F4WTA6 A0A1B6DS46 A0A1B6M6Z4 A0A0K2TAN0 Q7YSZ1 W8BKT3 V5GFQ0 A0A2P8ZJ36 T1HRF9 A0A0P4VQE4 B4KL07 B4Q3W0 Q9VII1 B4P720 A0A1I8MA47 Q71D96 A0A2P8ZDE0 B4HDE4 B4LUN2 A0A2J7QW68 A0A034WQS1 A0FD26 A0A1B6FV14 A0A067RDS9 A0A1E1WWY4 A0FD27 A0A1W4VFG9 A0A0M8ZZ44 B4JD92 A0FD25 A0A0A1XHA5 B4LSX1 B4LKT4 B4IFH6 W5J3F7
A0A0N1IH29 A0A1E1W402 S4P3Z0 A0A0V0G953 R4G540 A0A069DPK0 A0A224XN13 A0A0J9R4N1 A0A1I8M3Z3 B4MV65 B4IFH8 B4Q3W2 B5DIQ8 B4G719 A0A224XUK9 B4P719 A0A0V0G2Y8 A0A286P0W6 B4LUN1 T1IFT4 C5IWP3 A0A0P4VKI5 Q9VIH9 A0A1W4UVF0 A0A151JBA9 A0A023FD89 B4JD91 R4G7Y0 B3PAG6 A0A3B0K3I3 B3NKX1 A0A2P8YVI6 T1I2D3 E2J782 A0A1W4WPE1 E0VVY9 A0A0M4EEB1 A0A195EWX4 A0A067R4V8 B3MU77 R4UWG9 A0A067R6Z0 A0A182K0L5 A0A0L7R443 B3NKW9 A0A2J7QW69 A0A1S3D977 S4PL98 Q0MTF2 A0A195C5M5 B3NKX5 A0A182PPI2 B3PAG8 A0A182L1H1 A0NF51 A0A182HUP4 F4WTA6 A0A1B6DS46 A0A1B6M6Z4 A0A0K2TAN0 Q7YSZ1 W8BKT3 V5GFQ0 A0A2P8ZJ36 T1HRF9 A0A0P4VQE4 B4KL07 B4Q3W0 Q9VII1 B4P720 A0A1I8MA47 Q71D96 A0A2P8ZDE0 B4HDE4 B4LUN2 A0A2J7QW68 A0A034WQS1 A0FD26 A0A1B6FV14 A0A067RDS9 A0A1E1WWY4 A0FD27 A0A1W4VFG9 A0A0M8ZZ44 B4JD92 A0FD25 A0A0A1XHA5 B4LSX1 B4LKT4 B4IFH6 W5J3F7
Pubmed
19121390
28756777
22651552
26354079
23622113
26334808
+ More
22936249 25315136 17994087 15632085 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 29403074 20566863 24845553 20966253 12364791 14747013 17210077 21719571 14976983 24495485 14525923 25348373 17071113 28503490 25830018 20920257 23761445
22936249 25315136 17994087 15632085 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 29403074 20566863 24845553 20966253 12364791 14747013 17210077 21719571 14976983 24495485 14525923 25348373 17071113 28503490 25830018 20920257 23761445
EMBL
BABH01012989
NWSH01003451
PCG66274.1
KZ149910
PZC78145.1
AK402037
+ More
BAM18659.1 AK402651 BAM19273.1 KQ458756 KPJ05285.1 KQ460317 KPJ16013.1 GDQN01009348 JAT81706.1 GAIX01007881 JAA84679.1 GECL01001598 JAP04526.1 ACPB03025722 GAHY01000820 JAA76690.1 GBGD01003342 JAC85547.1 GFTR01002540 JAW13886.1 CM002910 KMY91237.1 CH963857 EDW76410.1 CH480833 EDW46400.1 CM000361 EDX05678.1 CH379060 EDY70200.1 CH479180 EDW29218.1 GFTR01002928 JAW13498.1 CM000158 EDW89988.1 GECL01003781 JAP02343.1 AB999705 BBA30659.1 CH940649 EDW64217.1 ACPB03001161 FJ968776 ACR56003.1 GDKW01003584 JAI53011.1 AE014134 AY119487 KX531372 AAF53940.1 AAM50141.1 AGB93181.1 ANY27182.1 KQ979147 KYN22441.1 GBBI01000023 JAC18689.1 CH916368 EDW03264.1 GAHY01001937 JAA75573.1 CH954451 EDV45278.2 OUUW01000004 SPP79591.1 CH954179 EDV54494.1 PYGN01000337 PSN48245.1 HP429300 ADN29800.1 DS235816 EEB17545.1 CP012523 ALC40325.1 ALC40335.1 KQ981940 KYN32728.1 KK852699 KDR18172.1 CH902624 EDV33406.1 KC740812 AGM32636.1 KDR18173.1 KQ414659 KOC65618.1 EDV54492.2 NEVH01009767 PNF32830.1 GAIX01004140 JAA88420.1 DQ846783 ABH09417.1 KQ978231 KYM96147.1 EDV54498.1 EDV45280.1 AAAB01008964 EAU76347.1 APCN01002469 GL888334 EGI62569.1 GEDC01008796 JAS28502.1 GEBQ01008282 JAT31695.1 HACA01005788 CDW23149.1 ACPB03009472 ACPB03009473 ACPB03009474 AY340271 GAHY01000792 AAQ20836.1 JAA76718.1 GAMC01007173 JAB99382.1 GALX01005657 JAB62809.1 PYGN01000040 PSN56518.1 ACPB03031755 GDKW01002540 JAI54055.1 CH933807 EDW12757.1 EDX05676.1 KMY91236.1 BT032654 AAF53938.1 ACD81668.1 EDW89989.1 AF531973 AAQ09872.1 PYGN01000090 PSN54515.1 CH480180 EDW26664.1 EDW64218.1 PNF32829.1 GAKP01002854 JAC56098.1 DQ907921 ABJ97669.1 GECZ01015723 JAS54046.1 KDR18170.1 GFAC01007653 JAT91535.1 DQ907922 ABJ97670.1 KQ435819 KOX72519.1 EDW03265.1 DQ907920 ABJ97668.1 GBXI01003523 JAD10769.1 EDW64882.1 CH940648 EDW60738.1 EDW46398.1 ADMH02002111 ETN58882.1
BAM18659.1 AK402651 BAM19273.1 KQ458756 KPJ05285.1 KQ460317 KPJ16013.1 GDQN01009348 JAT81706.1 GAIX01007881 JAA84679.1 GECL01001598 JAP04526.1 ACPB03025722 GAHY01000820 JAA76690.1 GBGD01003342 JAC85547.1 GFTR01002540 JAW13886.1 CM002910 KMY91237.1 CH963857 EDW76410.1 CH480833 EDW46400.1 CM000361 EDX05678.1 CH379060 EDY70200.1 CH479180 EDW29218.1 GFTR01002928 JAW13498.1 CM000158 EDW89988.1 GECL01003781 JAP02343.1 AB999705 BBA30659.1 CH940649 EDW64217.1 ACPB03001161 FJ968776 ACR56003.1 GDKW01003584 JAI53011.1 AE014134 AY119487 KX531372 AAF53940.1 AAM50141.1 AGB93181.1 ANY27182.1 KQ979147 KYN22441.1 GBBI01000023 JAC18689.1 CH916368 EDW03264.1 GAHY01001937 JAA75573.1 CH954451 EDV45278.2 OUUW01000004 SPP79591.1 CH954179 EDV54494.1 PYGN01000337 PSN48245.1 HP429300 ADN29800.1 DS235816 EEB17545.1 CP012523 ALC40325.1 ALC40335.1 KQ981940 KYN32728.1 KK852699 KDR18172.1 CH902624 EDV33406.1 KC740812 AGM32636.1 KDR18173.1 KQ414659 KOC65618.1 EDV54492.2 NEVH01009767 PNF32830.1 GAIX01004140 JAA88420.1 DQ846783 ABH09417.1 KQ978231 KYM96147.1 EDV54498.1 EDV45280.1 AAAB01008964 EAU76347.1 APCN01002469 GL888334 EGI62569.1 GEDC01008796 JAS28502.1 GEBQ01008282 JAT31695.1 HACA01005788 CDW23149.1 ACPB03009472 ACPB03009473 ACPB03009474 AY340271 GAHY01000792 AAQ20836.1 JAA76718.1 GAMC01007173 JAB99382.1 GALX01005657 JAB62809.1 PYGN01000040 PSN56518.1 ACPB03031755 GDKW01002540 JAI54055.1 CH933807 EDW12757.1 EDX05676.1 KMY91236.1 BT032654 AAF53938.1 ACD81668.1 EDW89989.1 AF531973 AAQ09872.1 PYGN01000090 PSN54515.1 CH480180 EDW26664.1 EDW64218.1 PNF32829.1 GAKP01002854 JAC56098.1 DQ907921 ABJ97669.1 GECZ01015723 JAS54046.1 KDR18170.1 GFAC01007653 JAT91535.1 DQ907922 ABJ97670.1 KQ435819 KOX72519.1 EDW03265.1 DQ907920 ABJ97668.1 GBXI01003523 JAD10769.1 EDW64882.1 CH940648 EDW60738.1 EDW46398.1 ADMH02002111 ETN58882.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000015103
UP000095301
+ More
UP000007798 UP000001292 UP000000304 UP000001819 UP000008744 UP000002282 UP000008792 UP000000803 UP000192221 UP000078492 UP000001070 UP000008711 UP000268350 UP000245037 UP000192223 UP000009046 UP000092553 UP000078541 UP000027135 UP000007801 UP000075881 UP000053825 UP000235965 UP000079169 UP000078542 UP000075885 UP000075882 UP000007062 UP000075840 UP000007755 UP000009192 UP000053105 UP000000673
UP000007798 UP000001292 UP000000304 UP000001819 UP000008744 UP000002282 UP000008792 UP000000803 UP000192221 UP000078492 UP000001070 UP000008711 UP000268350 UP000245037 UP000192223 UP000009046 UP000092553 UP000078541 UP000027135 UP000007801 UP000075881 UP000053825 UP000235965 UP000079169 UP000078542 UP000075885 UP000075882 UP000007062 UP000075840 UP000007755 UP000009192 UP000053105 UP000000673
Pfam
PF17064 QVR
Interpro
IPR031424
QVR
ProteinModelPortal
H9JH50
A0A2A4J434
A0A2W1BWX5
I4DL69
I4DMY3
A0A194QK48
+ More
A0A0N1IH29 A0A1E1W402 S4P3Z0 A0A0V0G953 R4G540 A0A069DPK0 A0A224XN13 A0A0J9R4N1 A0A1I8M3Z3 B4MV65 B4IFH8 B4Q3W2 B5DIQ8 B4G719 A0A224XUK9 B4P719 A0A0V0G2Y8 A0A286P0W6 B4LUN1 T1IFT4 C5IWP3 A0A0P4VKI5 Q9VIH9 A0A1W4UVF0 A0A151JBA9 A0A023FD89 B4JD91 R4G7Y0 B3PAG6 A0A3B0K3I3 B3NKX1 A0A2P8YVI6 T1I2D3 E2J782 A0A1W4WPE1 E0VVY9 A0A0M4EEB1 A0A195EWX4 A0A067R4V8 B3MU77 R4UWG9 A0A067R6Z0 A0A182K0L5 A0A0L7R443 B3NKW9 A0A2J7QW69 A0A1S3D977 S4PL98 Q0MTF2 A0A195C5M5 B3NKX5 A0A182PPI2 B3PAG8 A0A182L1H1 A0NF51 A0A182HUP4 F4WTA6 A0A1B6DS46 A0A1B6M6Z4 A0A0K2TAN0 Q7YSZ1 W8BKT3 V5GFQ0 A0A2P8ZJ36 T1HRF9 A0A0P4VQE4 B4KL07 B4Q3W0 Q9VII1 B4P720 A0A1I8MA47 Q71D96 A0A2P8ZDE0 B4HDE4 B4LUN2 A0A2J7QW68 A0A034WQS1 A0FD26 A0A1B6FV14 A0A067RDS9 A0A1E1WWY4 A0FD27 A0A1W4VFG9 A0A0M8ZZ44 B4JD92 A0FD25 A0A0A1XHA5 B4LSX1 B4LKT4 B4IFH6 W5J3F7
A0A0N1IH29 A0A1E1W402 S4P3Z0 A0A0V0G953 R4G540 A0A069DPK0 A0A224XN13 A0A0J9R4N1 A0A1I8M3Z3 B4MV65 B4IFH8 B4Q3W2 B5DIQ8 B4G719 A0A224XUK9 B4P719 A0A0V0G2Y8 A0A286P0W6 B4LUN1 T1IFT4 C5IWP3 A0A0P4VKI5 Q9VIH9 A0A1W4UVF0 A0A151JBA9 A0A023FD89 B4JD91 R4G7Y0 B3PAG6 A0A3B0K3I3 B3NKX1 A0A2P8YVI6 T1I2D3 E2J782 A0A1W4WPE1 E0VVY9 A0A0M4EEB1 A0A195EWX4 A0A067R4V8 B3MU77 R4UWG9 A0A067R6Z0 A0A182K0L5 A0A0L7R443 B3NKW9 A0A2J7QW69 A0A1S3D977 S4PL98 Q0MTF2 A0A195C5M5 B3NKX5 A0A182PPI2 B3PAG8 A0A182L1H1 A0NF51 A0A182HUP4 F4WTA6 A0A1B6DS46 A0A1B6M6Z4 A0A0K2TAN0 Q7YSZ1 W8BKT3 V5GFQ0 A0A2P8ZJ36 T1HRF9 A0A0P4VQE4 B4KL07 B4Q3W0 Q9VII1 B4P720 A0A1I8MA47 Q71D96 A0A2P8ZDE0 B4HDE4 B4LUN2 A0A2J7QW68 A0A034WQS1 A0FD26 A0A1B6FV14 A0A067RDS9 A0A1E1WWY4 A0FD27 A0A1W4VFG9 A0A0M8ZZ44 B4JD92 A0FD25 A0A0A1XHA5 B4LSX1 B4LKT4 B4IFH6 W5J3F7
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 21,
Likelihood: 0.998686
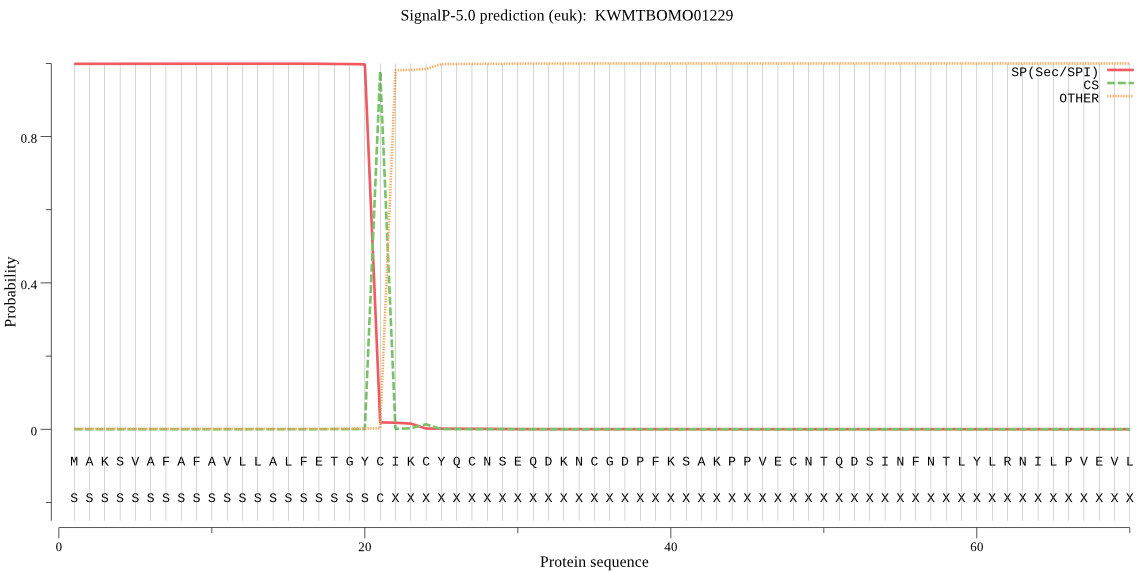
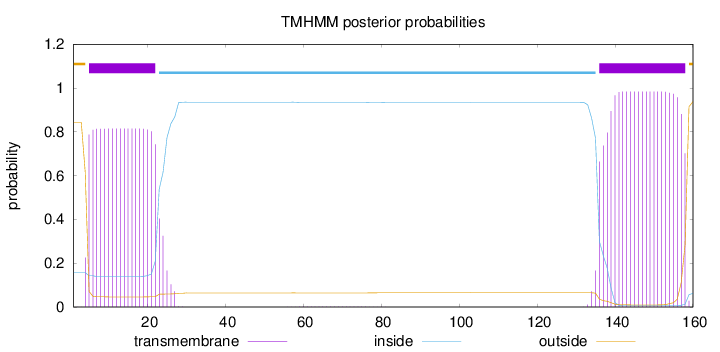
Length:
160
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
37.49227
Exp number, first 60 AAs:
15.84516
Total prob of N-in:
0.15759
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 22
inside
23 - 135
TMhelix
136 - 158
outside
159 - 160
Population Genetic Test Statistics
Pi
311.614393
Theta
182.148178
Tajima's D
1.472253
CLR
0.537927
CSRT
0.781510924453777
Interpretation
Uncertain
