Gene
KWMTBOMO01226 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008846
Annotation
PREDICTED:_NFU1_iron-sulfur_cluster_scaffold_homolog?_mitochondrial-like_[Amyelois_transitella]
Full name
XK-related protein
+ More
Serine/threonine-protein phosphatase
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial
Serine/threonine-protein phosphatase
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial
Location in the cell
Cytoplasmic Reliability : 2.23
Sequence
CDS
ATGTTCATACAAACTCAAGACACACCAAATCCAAATAGCTTGAAGTTTTTACCTGGTACTAAAGTTCTGGAGCCGGGTCAAACTTTGGACTTTCCTAATATTGGAGCTGCACACTGCAGTCCACTTGCCAAAATGTTATTCCGCATTGATGGAGTGAAAGCAGTCTTCTTTGGACCAGAATTCATTACTGTAACAAAACAGGACGATGATGTTGATTGGAAGCTATTAAAACCTGAAATATTTGGTACAATAATGGATTTCTTTGCCAGCGGACTGCCTGTGGTAACAGATGCTAAACCTTCGGGTGATACTCAGATAAATGAAGATGATGATGAAATAGTTCAAATGATAAAAGAATTGCTGGACACAAGGATTCGTCCTACTGTACAAGAAGATGGTGGCGATGTACTGTTTGTTGATTTTAAAGATGGAATCCTGAGGCTAAAGATGCAAGGATCGTGCTCTTCTTGTCCAAGCTCTATTGTAACTTTAAAAAATGGGGTACAGAATATGATGCAGTTTTACATACCAGAAGTTTTGGCTGTCGAACAAATAGACGATGAAGGCGAAAAGCTAAGCGAAAAAATCTTCCGGGAATTCGAGCAAAGTAAAGCCAAGGATGCAAACAAAGATACATAA
Protein
MFIQTQDTPNPNSLKFLPGTKVLEPGQTLDFPNIGAAHCSPLAKMLFRIDGVKAVFFGPEFITVTKQDDDVDWKLLKPEIFGTIMDFFASGLPVVTDAKPSGDTQINEDDDEIVQMIKELLDTRIRPTVQEDGGDVLFVDFKDGILRLKMQGSCSSCPSSIVTLKNGVQNMMQFYIPEVLAVEQIDDEGEKLSEKIFREFEQSKAKDANKDT
Summary
Description
Molecular scaffold for [Fe-S] cluster assembly of mitochondrial iron-sulfur proteins.
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Similarity
Belongs to the XK family.
Belongs to the PPP phosphatase family.
Belongs to the NifU family.
Belongs to the PPP phosphatase family.
Belongs to the NifU family.
Keywords
Complete proteome
Iron
Iron-sulfur
Metal-binding
Mitochondrion
Reference proteome
Transit peptide
Feature
chain NFU1 iron-sulfur cluster scaffold homolog, mitochondrial
Uniprot
H9JH49
A0A194QIE5
A0A2A4J1M5
A0A2W1BT24
A0A2H1W6S2
A0A0N1IAN2
+ More
A0A1B6KTI5 A0A0C9QK14 A0A310SPM7 A0A1B6C2S0 A0A212FF20 A0A1Y1L4S8 A0A1B6LTF8 A0A0L7R3C4 A0A182TQN0 A0A182IA40 A0A182W4D9 A0A182WW50 A0A026WMJ3 A0A088A2E5 A0A182Y7M5 D6WV84 A0A158NW89 A0A182VHC1 Q7QFP2 A0A1W4WS11 A0A182M639 A0A1W4WRR7 A0A084VV33 A0A1W4WQT1 A0A182RZ52 A0A182KJ99 A0A182N059 A0A1Q3G4D7 A0A1Q3G4D6 A0A182PZJ4 E9IG51 A0A2H8TWQ1 A0A195FSN4 A0A182JVS9 A0A0A9XKZ0 A0A182P1N7 Q17J52 C4WRP3 A0A1Q3G4D5 A0A023F486 A0A182JEA0 J9JZG9 A0A2J7QGN9 A0A023ENN5 A0A2J7QGP4 A0A195C4Z2 A0A067R6Z7 A0A195DIA5 F4WBU5 A0A3Q0IVK2 A0A0P4VTP8 E2B2K2 A0A151WN79 A0A2M4AJ47 A0A182G518 A0A2M4AJ34 T1IAX6 A0A0V0G9D9 W5JT37 A0A182HBT4 A0A069DRU6 T1E8K6 A0A224XW79 A0A182FLN4 A0A2S2QF64 K1QY89 A0A195BE99 A0A0K8TTC9 E1ZX96 A0A210Q9S0 A0A0L7LPF5 A0A2I6UI50 A0A2R7W596 K7IXB3 A0A023ELN7 A0A2C9JLK5 T1PCY1 N6TIY4 A0A161MLG8 A0A0T6AVY0 A0A1J1J5V1 B4JWR9 B4M375 A0A0B7B7S8 B4NE93 C1BQS4 A0A0A1WRK4 B4L310 A0A1B0BIV5 A0A1A9Y8R6 A0A0M5J6A8 A0A2T7PZ18 A0A240SWW9 B4H303
A0A1B6KTI5 A0A0C9QK14 A0A310SPM7 A0A1B6C2S0 A0A212FF20 A0A1Y1L4S8 A0A1B6LTF8 A0A0L7R3C4 A0A182TQN0 A0A182IA40 A0A182W4D9 A0A182WW50 A0A026WMJ3 A0A088A2E5 A0A182Y7M5 D6WV84 A0A158NW89 A0A182VHC1 Q7QFP2 A0A1W4WS11 A0A182M639 A0A1W4WRR7 A0A084VV33 A0A1W4WQT1 A0A182RZ52 A0A182KJ99 A0A182N059 A0A1Q3G4D7 A0A1Q3G4D6 A0A182PZJ4 E9IG51 A0A2H8TWQ1 A0A195FSN4 A0A182JVS9 A0A0A9XKZ0 A0A182P1N7 Q17J52 C4WRP3 A0A1Q3G4D5 A0A023F486 A0A182JEA0 J9JZG9 A0A2J7QGN9 A0A023ENN5 A0A2J7QGP4 A0A195C4Z2 A0A067R6Z7 A0A195DIA5 F4WBU5 A0A3Q0IVK2 A0A0P4VTP8 E2B2K2 A0A151WN79 A0A2M4AJ47 A0A182G518 A0A2M4AJ34 T1IAX6 A0A0V0G9D9 W5JT37 A0A182HBT4 A0A069DRU6 T1E8K6 A0A224XW79 A0A182FLN4 A0A2S2QF64 K1QY89 A0A195BE99 A0A0K8TTC9 E1ZX96 A0A210Q9S0 A0A0L7LPF5 A0A2I6UI50 A0A2R7W596 K7IXB3 A0A023ELN7 A0A2C9JLK5 T1PCY1 N6TIY4 A0A161MLG8 A0A0T6AVY0 A0A1J1J5V1 B4JWR9 B4M375 A0A0B7B7S8 B4NE93 C1BQS4 A0A0A1WRK4 B4L310 A0A1B0BIV5 A0A1A9Y8R6 A0A0M5J6A8 A0A2T7PZ18 A0A240SWW9 B4H303
EC Number
3.1.3.16
Pubmed
19121390
26354079
28756777
22118469
28004739
24508170
+ More
30249741 25244985 18362917 19820115 21347285 12364791 14747013 17210077 24438588 20966253 21282665 25401762 26823975 17510324 25474469 24945155 24845553 21719571 27129103 20798317 26483478 20920257 23761445 26334808 22992520 26369729 28812685 26227816 29326599 20075255 15562597 25315136 23537049 17994087 25830018
30249741 25244985 18362917 19820115 21347285 12364791 14747013 17210077 24438588 20966253 21282665 25401762 26823975 17510324 25474469 24945155 24845553 21719571 27129103 20798317 26483478 20920257 23761445 26334808 22992520 26369729 28812685 26227816 29326599 20075255 15562597 25315136 23537049 17994087 25830018
EMBL
BABH01012993
KQ458756
KPJ05287.1
NWSH01004274
PCG65283.1
KZ150022
+ More
PZC74873.1 ODYU01006706 SOQ48795.1 KQ460317 KPJ16015.1 GEBQ01031006 GEBQ01025227 JAT08971.1 JAT14750.1 GBYB01015148 JAG84915.1 KQ761976 OAD56422.1 GEDC01029486 JAS07812.1 AGBW02008884 OWR52303.1 GEZM01064600 JAV68699.1 GEBQ01013022 JAT26955.1 KQ414663 KOC65375.1 APCN01001102 KK107154 QOIP01000001 EZA56886.1 RLU27537.1 KQ971363 EFA07765.1 ADTU01027850 AAAB01008846 EAA06366.5 AXCM01001777 ATLV01017149 KE525157 KFB41827.1 GFDL01000370 JAV34675.1 GFDL01000382 JAV34663.1 AXCN02001308 GL762910 EFZ20436.1 GFXV01006920 MBW18725.1 KQ981280 KYN43456.1 GBHO01023283 GBRD01011598 GDHC01013043 GDHC01006661 JAG20321.1 JAG54226.1 JAQ05586.1 JAQ11968.1 CH477235 EAT46657.1 AK339854 BAH70563.1 GFDL01000373 JAV34672.1 GBBI01002382 JAC16330.1 ABLF02027172 NEVH01014361 PNF27749.1 GAPW01003559 JAC10039.1 PNF27751.1 KQ978317 KYM95228.1 KK852699 KDR18183.1 KQ980824 KYN12567.1 GL888066 EGI68373.1 GDKW01003361 JAI53234.1 GL445147 EFN90079.1 KQ982911 KYQ49324.1 GGFK01007446 MBW40767.1 JXUM01042847 KQ561341 KXJ78907.1 GGFK01007479 MBW40800.1 ACPB03005351 GECL01001483 JAP04641.1 ADMH02000225 ETN67306.1 JXUM01125673 KQ566972 KXJ69693.1 GBGD01002483 JAC86406.1 GAMD01002683 JAA98907.1 GFTR01004215 JAW12211.1 GGMS01007175 MBY76378.1 JH816692 EKC36093.1 EKC36094.1 KQ976511 KYM82522.1 GDAI01000413 JAI17190.1 GL435030 EFN74186.1 NEDP02004497 OWF45476.1 JTDY01000411 KOB77317.1 MG596304 AUO79641.1 KK854228 PTY13385.1 AAZX01011471 GAPW01003425 JAC10173.1 KA646647 AFP61276.1 APGK01036009 KB740932 KB632408 ENN77798.1 ERL95147.1 GEMB01002345 JAS00838.1 LJIG01022688 KRT79238.1 CVRI01000070 CRL07370.1 CH916376 EDV95195.1 CH940651 EDW65250.1 HACG01042188 CEK89053.1 CH964239 EDW82062.1 BT076953 ACO11377.1 GBXI01013147 JAD01145.1 CH933810 EDW07896.2 JXJN01015182 CP012528 ALC49468.1 PZQS01000001 PVD38658.1 CCAG010017550 CH479205 EDW30835.1
PZC74873.1 ODYU01006706 SOQ48795.1 KQ460317 KPJ16015.1 GEBQ01031006 GEBQ01025227 JAT08971.1 JAT14750.1 GBYB01015148 JAG84915.1 KQ761976 OAD56422.1 GEDC01029486 JAS07812.1 AGBW02008884 OWR52303.1 GEZM01064600 JAV68699.1 GEBQ01013022 JAT26955.1 KQ414663 KOC65375.1 APCN01001102 KK107154 QOIP01000001 EZA56886.1 RLU27537.1 KQ971363 EFA07765.1 ADTU01027850 AAAB01008846 EAA06366.5 AXCM01001777 ATLV01017149 KE525157 KFB41827.1 GFDL01000370 JAV34675.1 GFDL01000382 JAV34663.1 AXCN02001308 GL762910 EFZ20436.1 GFXV01006920 MBW18725.1 KQ981280 KYN43456.1 GBHO01023283 GBRD01011598 GDHC01013043 GDHC01006661 JAG20321.1 JAG54226.1 JAQ05586.1 JAQ11968.1 CH477235 EAT46657.1 AK339854 BAH70563.1 GFDL01000373 JAV34672.1 GBBI01002382 JAC16330.1 ABLF02027172 NEVH01014361 PNF27749.1 GAPW01003559 JAC10039.1 PNF27751.1 KQ978317 KYM95228.1 KK852699 KDR18183.1 KQ980824 KYN12567.1 GL888066 EGI68373.1 GDKW01003361 JAI53234.1 GL445147 EFN90079.1 KQ982911 KYQ49324.1 GGFK01007446 MBW40767.1 JXUM01042847 KQ561341 KXJ78907.1 GGFK01007479 MBW40800.1 ACPB03005351 GECL01001483 JAP04641.1 ADMH02000225 ETN67306.1 JXUM01125673 KQ566972 KXJ69693.1 GBGD01002483 JAC86406.1 GAMD01002683 JAA98907.1 GFTR01004215 JAW12211.1 GGMS01007175 MBY76378.1 JH816692 EKC36093.1 EKC36094.1 KQ976511 KYM82522.1 GDAI01000413 JAI17190.1 GL435030 EFN74186.1 NEDP02004497 OWF45476.1 JTDY01000411 KOB77317.1 MG596304 AUO79641.1 KK854228 PTY13385.1 AAZX01011471 GAPW01003425 JAC10173.1 KA646647 AFP61276.1 APGK01036009 KB740932 KB632408 ENN77798.1 ERL95147.1 GEMB01002345 JAS00838.1 LJIG01022688 KRT79238.1 CVRI01000070 CRL07370.1 CH916376 EDV95195.1 CH940651 EDW65250.1 HACG01042188 CEK89053.1 CH964239 EDW82062.1 BT076953 ACO11377.1 GBXI01013147 JAD01145.1 CH933810 EDW07896.2 JXJN01015182 CP012528 ALC49468.1 PZQS01000001 PVD38658.1 CCAG010017550 CH479205 EDW30835.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000053825
+ More
UP000075902 UP000075840 UP000075920 UP000076407 UP000053097 UP000279307 UP000005203 UP000076408 UP000007266 UP000005205 UP000075903 UP000007062 UP000192223 UP000075883 UP000030765 UP000075900 UP000075882 UP000075884 UP000075886 UP000078541 UP000075881 UP000075885 UP000008820 UP000075880 UP000007819 UP000235965 UP000078542 UP000027135 UP000078492 UP000007755 UP000079169 UP000008237 UP000075809 UP000069940 UP000249989 UP000015103 UP000000673 UP000069272 UP000005408 UP000078540 UP000000311 UP000242188 UP000037510 UP000002358 UP000076420 UP000095301 UP000019118 UP000030742 UP000183832 UP000001070 UP000008792 UP000007798 UP000009192 UP000092460 UP000092443 UP000092553 UP000245119 UP000092444 UP000008744
UP000075902 UP000075840 UP000075920 UP000076407 UP000053097 UP000279307 UP000005203 UP000076408 UP000007266 UP000005205 UP000075903 UP000007062 UP000192223 UP000075883 UP000030765 UP000075900 UP000075882 UP000075884 UP000075886 UP000078541 UP000075881 UP000075885 UP000008820 UP000075880 UP000007819 UP000235965 UP000078542 UP000027135 UP000078492 UP000007755 UP000079169 UP000008237 UP000075809 UP000069940 UP000249989 UP000015103 UP000000673 UP000069272 UP000005408 UP000078540 UP000000311 UP000242188 UP000037510 UP000002358 UP000076420 UP000095301 UP000019118 UP000030742 UP000183832 UP000001070 UP000008792 UP000007798 UP000009192 UP000092460 UP000092443 UP000092553 UP000245119 UP000092444 UP000008744
Interpro
IPR001075
NIF_FeS_clus_asmbl_NifU_C
+ More
IPR036498 Nfu/NifU_N_sf
IPR034904 FSCA_dom_sf
IPR014824 Nfu/NifU_N
IPR035433 NFU1-like
IPR036085 PAZ_dom_sf
IPR036397 RNaseH_sf
IPR003165 Piwi
IPR012337 RNaseH-like_sf
IPR018629 XK-rel
IPR004843 Calcineurin-like_PHP_ApaH
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR029052 Metallo-depent_PP-like
IPR036498 Nfu/NifU_N_sf
IPR034904 FSCA_dom_sf
IPR014824 Nfu/NifU_N
IPR035433 NFU1-like
IPR036085 PAZ_dom_sf
IPR036397 RNaseH_sf
IPR003165 Piwi
IPR012337 RNaseH-like_sf
IPR018629 XK-rel
IPR004843 Calcineurin-like_PHP_ApaH
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR029052 Metallo-depent_PP-like
ProteinModelPortal
H9JH49
A0A194QIE5
A0A2A4J1M5
A0A2W1BT24
A0A2H1W6S2
A0A0N1IAN2
+ More
A0A1B6KTI5 A0A0C9QK14 A0A310SPM7 A0A1B6C2S0 A0A212FF20 A0A1Y1L4S8 A0A1B6LTF8 A0A0L7R3C4 A0A182TQN0 A0A182IA40 A0A182W4D9 A0A182WW50 A0A026WMJ3 A0A088A2E5 A0A182Y7M5 D6WV84 A0A158NW89 A0A182VHC1 Q7QFP2 A0A1W4WS11 A0A182M639 A0A1W4WRR7 A0A084VV33 A0A1W4WQT1 A0A182RZ52 A0A182KJ99 A0A182N059 A0A1Q3G4D7 A0A1Q3G4D6 A0A182PZJ4 E9IG51 A0A2H8TWQ1 A0A195FSN4 A0A182JVS9 A0A0A9XKZ0 A0A182P1N7 Q17J52 C4WRP3 A0A1Q3G4D5 A0A023F486 A0A182JEA0 J9JZG9 A0A2J7QGN9 A0A023ENN5 A0A2J7QGP4 A0A195C4Z2 A0A067R6Z7 A0A195DIA5 F4WBU5 A0A3Q0IVK2 A0A0P4VTP8 E2B2K2 A0A151WN79 A0A2M4AJ47 A0A182G518 A0A2M4AJ34 T1IAX6 A0A0V0G9D9 W5JT37 A0A182HBT4 A0A069DRU6 T1E8K6 A0A224XW79 A0A182FLN4 A0A2S2QF64 K1QY89 A0A195BE99 A0A0K8TTC9 E1ZX96 A0A210Q9S0 A0A0L7LPF5 A0A2I6UI50 A0A2R7W596 K7IXB3 A0A023ELN7 A0A2C9JLK5 T1PCY1 N6TIY4 A0A161MLG8 A0A0T6AVY0 A0A1J1J5V1 B4JWR9 B4M375 A0A0B7B7S8 B4NE93 C1BQS4 A0A0A1WRK4 B4L310 A0A1B0BIV5 A0A1A9Y8R6 A0A0M5J6A8 A0A2T7PZ18 A0A240SWW9 B4H303
A0A1B6KTI5 A0A0C9QK14 A0A310SPM7 A0A1B6C2S0 A0A212FF20 A0A1Y1L4S8 A0A1B6LTF8 A0A0L7R3C4 A0A182TQN0 A0A182IA40 A0A182W4D9 A0A182WW50 A0A026WMJ3 A0A088A2E5 A0A182Y7M5 D6WV84 A0A158NW89 A0A182VHC1 Q7QFP2 A0A1W4WS11 A0A182M639 A0A1W4WRR7 A0A084VV33 A0A1W4WQT1 A0A182RZ52 A0A182KJ99 A0A182N059 A0A1Q3G4D7 A0A1Q3G4D6 A0A182PZJ4 E9IG51 A0A2H8TWQ1 A0A195FSN4 A0A182JVS9 A0A0A9XKZ0 A0A182P1N7 Q17J52 C4WRP3 A0A1Q3G4D5 A0A023F486 A0A182JEA0 J9JZG9 A0A2J7QGN9 A0A023ENN5 A0A2J7QGP4 A0A195C4Z2 A0A067R6Z7 A0A195DIA5 F4WBU5 A0A3Q0IVK2 A0A0P4VTP8 E2B2K2 A0A151WN79 A0A2M4AJ47 A0A182G518 A0A2M4AJ34 T1IAX6 A0A0V0G9D9 W5JT37 A0A182HBT4 A0A069DRU6 T1E8K6 A0A224XW79 A0A182FLN4 A0A2S2QF64 K1QY89 A0A195BE99 A0A0K8TTC9 E1ZX96 A0A210Q9S0 A0A0L7LPF5 A0A2I6UI50 A0A2R7W596 K7IXB3 A0A023ELN7 A0A2C9JLK5 T1PCY1 N6TIY4 A0A161MLG8 A0A0T6AVY0 A0A1J1J5V1 B4JWR9 B4M375 A0A0B7B7S8 B4NE93 C1BQS4 A0A0A1WRK4 B4L310 A0A1B0BIV5 A0A1A9Y8R6 A0A0M5J6A8 A0A2T7PZ18 A0A240SWW9 B4H303
PDB
2LTM
E-value=2.67199e-32,
Score=343
Ontologies
GO
Topology
Subcellular location
Membrane
Mitochondrion
Mitochondrion
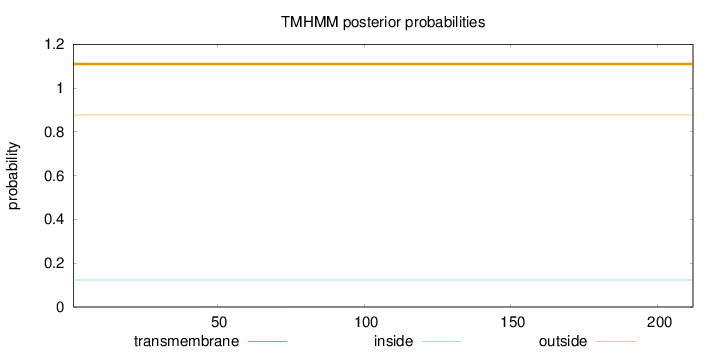
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00903999999999998
Exp number, first 60 AAs:
0.00237
Total prob of N-in:
0.12347
outside
1 - 212
Population Genetic Test Statistics
Pi
377.919092
Theta
215.240239
Tajima's D
2.030812
CLR
0.106637
CSRT
0.890955452227389
Interpretation
Uncertain
