Gene
KWMTBOMO01214
Pre Gene Modal
BGIBMGA014548
Annotation
PREDICTED:_MOXD1_homolog_1-like_[Plutella_xylostella]
Full name
MOXD1 homolog 1
Location in the cell
Cytoplasmic Reliability : 2.292
Sequence
CDS
ATGATAGGAGAGAGTGCTGGAGAGGGAGAACGCGGAGGGGTTCTCAACCTAGGAGCATCTACCGTCCTAGGGAGTGCCCTAGGTTTGGGGATGGGTTTCTTGGGTGAGGGAAGGGATTCCTTTGAGGGGGACGTTGAGCGAGAGGAGGAGGGGGAGGACGAAGCAGATGTGGATGCAGAAGAGGAAGGAGTGACGGAGGTAGCTTCGCTATCGTCACCCTCGGATCTAGAAGAGGGGCCTATCCTCGGTCTCTTGTCTTTCTCCTCAGAAGAGCCCCGGTCGGACTCGGACCTGGCCTTCTTTCGCGGCCGAGGGGTCCCCGAGTTATCGGTCGGTAAGGGAAAGCAACTTTCCCCCGAACCGATTCTCGGCGGTCCCTGGGCCGCTCCAGAAGGCCCGGGCTTATTGTCCGGGCCTGGGCGAGTTTGGGTTTGGGCCAATAGGCTAGTTCCTATTGATTGTCACGGACCCGAGCACGAAGATCTACCAGTAGCCGACGACGTTCAGAATTACGAGCTCATATCTGGGTATCAGAATGCGACCCACACGACTGTGGAATTTCGGAGGCAGCTCAACACTTGCGATAGACAAGACTTTGTTATTGGCGAGGACACAGTACAAGTGCTTTGGGCGCTTGGACCTGAGGGCTCAGATGGACAGCTTCCAAAACACATGAAGAGTGGATCTAGACCATTAAGACTTCTGCAACCAGTGTCGAAACCCGAGACGATACCTCTAAAACATTGGGATGTCAGATTGACTAATTTGACCATCCCCCACGATATGGCAACGCTATTTTGGTGTAAAATCTTCAAAGCTCCGGACTTGCCTAAGAAGCATCATATAATTGGCTACCAACCTCTGATCGATAGTCGGCCAATAAAGAACGGCAAACCCGTCGCAGACAATAATAGTTTTTCGCCTGTACATCATATGGTTCTATATGAATGCGCTGAAGACGAAGACAAACACCTGTGGAACGTTTGGGCTGATAGCGACGGGTATTTTGGACCAATACGGCCAAGTGGTTGGGCTACTTGTGCCAGGCCAATTGCGGCTTGGGCTATTGGATCTCAAGGTGAATTCCTGCCGGAAAGCGTCGGTATACCCTTGGGGGAAGACGGAGGGGTTTCGTTCTACATGCTCGAGGTTCATTACGATAATCAAGCGTTGCACGACGTTCTGGACAGTTCCGGTATAAGGGTGCACTACACGCCCGCTTTGAGGCAACACGACGCCGCCCTCCTCGGCGCCGGCATAGGGGTGTCGGCCCTCCACGTGATCCCCCCGAAACAACGGCGCTACAGGACCGTGGGGATATGCAGCTCCGAGTGTACCGACACGACCCTACCCGACGAGGGCATCAACATCGTGTCAGTTCTTCTTCACGCTCACGGAACAGCGAGAAAGATAGCCCTCAAACATGTTAGGGGCACGCAGGAGTTGCCGAGGATATCGGAGGAGAACTCATATGATGCTCGCTACCAACAATCAAGAGTGGTGCCCGGCGGTAGGAAATTCAAGAGAGGAGATACGCTGATCACTGAATGCACTTACGACACTACGTCGAGAACTAAACCTATTTTGGGGGGCTATTCTGCGAAACAGGAAATGTGCCTATCCTTCATTTTGTACTACCCTCGTACGGAGCTGGCGGGTTGCTATTCAATGACACCGGTCAAAGAGTTCTTCGAAACGTTCGGAGTCAAAAAGTTTTACGGATTGAACTTTACGCAAGTCGAAAACATATTTTTGTCATCCGGAAGCCTCGAAAATTTACCGCCGCAGCTGGAGATCGACGTGAAACAGAGGAGCACCGACGAAACCGCCAAGCAAGGAGACGATCAAGGCGTTGGACTGATGAAGGAACTAGTGATCGTTGAACCGTCGGAGTTCCTGAACAAGTCGTTCATGGATCACCTCAACGAGATGCCCTGGGAGGAGCCGCTGCTGACCGAACAGATCGAGAAGTCCCTGTACAACGGCATGCACATGACCTTCTGCAAGAAACGGGACGATTCCTGGGGAATGCCCATACAAATACAGAACTACCCTGCGTATTCGGAATTAACGACCAACGAAGCGTCCGAAAAATCTTGCAACTACAAGAGTGTGAGACTACCCTCTGTAACGAAGTCTTCGACGGGCGCGGCCGCACATACGACTTTAAGCTCGATCACACTGCTCGTTTGCATCTCGCTTGCGGCGACGCGGTAA
Protein
MIGESAGEGERGGVLNLGASTVLGSALGLGMGFLGEGRDSFEGDVEREEEGEDEADVDAEEEGVTEVASLSSPSDLEEGPILGLLSFSSEEPRSDSDLAFFRGRGVPELSVGKGKQLSPEPILGGPWAAPEGPGLLSGPGRVWVWANRLVPIDCHGPEHEDLPVADDVQNYELISGYQNATHTTVEFRRQLNTCDRQDFVIGEDTVQVLWALGPEGSDGQLPKHMKSGSRPLRLLQPVSKPETIPLKHWDVRLTNLTIPHDMATLFWCKIFKAPDLPKKHHIIGYQPLIDSRPIKNGKPVADNNSFSPVHHMVLYECAEDEDKHLWNVWADSDGYFGPIRPSGWATCARPIAAWAIGSQGEFLPESVGIPLGEDGGVSFYMLEVHYDNQALHDVLDSSGIRVHYTPALRQHDAALLGAGIGVSALHVIPPKQRRYRTVGICSSECTDTTLPDEGINIVSVLLHAHGTARKIALKHVRGTQELPRISEENSYDARYQQSRVVPGGRKFKRGDTLITECTYDTTSRTKPILGGYSAKQEMCLSFILYYPRTELAGCYSMTPVKEFFETFGVKKFYGLNFTQVENIFLSSGSLENLPPQLEIDVKQRSTDETAKQGDDQGVGLMKELVIVEPSEFLNKSFMDHLNEMPWEEPLLTEQIEKSLYNGMHMTFCKKRDDSWGMPIQIQNYPAYSELTTNEASEKSCNYKSVRLPSVTKSSTGAAAHTTLSSITLLVCISLAATR
Summary
Cofactor
Cu(2+)
Similarity
Belongs to the copper type II ascorbate-dependent monooxygenase family.
Keywords
Complete proteome
Copper
Disulfide bond
Glycoprotein
Metal-binding
Monooxygenase
Oxidoreductase
Reference proteome
Secreted
Signal
Feature
chain MOXD1 homolog 1
Uniprot
A0A2A4J6Q1
A0A212FDE6
A0A194Q0W7
A0A212EHD6
B4KLU6
Q7Q325
+ More
A0A182VED0 A0A182U400 A0A182QCH8 A0A1I8M0I8 A0A182LPJ1 B4J6F9 A0A1I8M0I2 B4LPE5 A0A1I8PU30 A0A182XCB0 A0A182J9H7 A0A139WCR2 A0A182YDD4 A0A0L0CIG9 A0A0A1X7P4 A0A139WCN5 A0A034WHH5 A0A0K8U160 A0A1B0BT02 B4MQU2 A0A182P9D6 A0A084WBF4 A0A182WF50 A0A182FTC9 A0A336L841 A0A182HNL2 A0A0J7NWM9 A0A182RFB3 B4H8A3 A0A0C9Q5J7 B4QLT9 Q29CS1 A0A1A9ZYJ1 A0A1Y1KNQ2 A0A1B0G0W8 A0A3B0KSR7 M9PFT8 Q9VUY0 A0A2J7Q985 A0A1W4XC79 U4UCX0 N6TBP6 E9IPD3 B4PD26 A0A1W4W1E6 B3NDR5 E2AUK9 A0A154P125 A0A182GK54 A0A1B6C5X6 A0A067QW71 A0A0L7RD13 A0A3L8E3L6 A0A158P319 K7IPK7 A0A2H8TE63 A0A310SGK6 A0A0N0U712 A0A026WV84 A0A195BIF4 A0A2A3E7L9 A0A195EYX4 A0A232EJS7 A0A195CD26 A0A2P8YVT5 A0A2S2Q6E1 A0A195DN93 A0A1A9V9R5 A0A151WYR9 A0A1J1I6P5 A0A182K240 J9K956 A0A2S2PV31 A0A0P4W3V5 A0A0P4WP84 A0A0P4WAE3 A0A0P4WAC2 A0A2P2I4Z1 A0A1D2NGQ1 E0VDU7
A0A182VED0 A0A182U400 A0A182QCH8 A0A1I8M0I8 A0A182LPJ1 B4J6F9 A0A1I8M0I2 B4LPE5 A0A1I8PU30 A0A182XCB0 A0A182J9H7 A0A139WCR2 A0A182YDD4 A0A0L0CIG9 A0A0A1X7P4 A0A139WCN5 A0A034WHH5 A0A0K8U160 A0A1B0BT02 B4MQU2 A0A182P9D6 A0A084WBF4 A0A182WF50 A0A182FTC9 A0A336L841 A0A182HNL2 A0A0J7NWM9 A0A182RFB3 B4H8A3 A0A0C9Q5J7 B4QLT9 Q29CS1 A0A1A9ZYJ1 A0A1Y1KNQ2 A0A1B0G0W8 A0A3B0KSR7 M9PFT8 Q9VUY0 A0A2J7Q985 A0A1W4XC79 U4UCX0 N6TBP6 E9IPD3 B4PD26 A0A1W4W1E6 B3NDR5 E2AUK9 A0A154P125 A0A182GK54 A0A1B6C5X6 A0A067QW71 A0A0L7RD13 A0A3L8E3L6 A0A158P319 K7IPK7 A0A2H8TE63 A0A310SGK6 A0A0N0U712 A0A026WV84 A0A195BIF4 A0A2A3E7L9 A0A195EYX4 A0A232EJS7 A0A195CD26 A0A2P8YVT5 A0A2S2Q6E1 A0A195DN93 A0A1A9V9R5 A0A151WYR9 A0A1J1I6P5 A0A182K240 J9K956 A0A2S2PV31 A0A0P4W3V5 A0A0P4WP84 A0A0P4WAE3 A0A0P4WAC2 A0A2P2I4Z1 A0A1D2NGQ1 E0VDU7
EC Number
1.14.17.-
Pubmed
22118469
26354079
17994087
12364791
25315136
20966253
+ More
18362917 19820115 25244985 26108605 25830018 25348373 24438588 22936249 15632085 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 11900973 23537049 21282665 17550304 20798317 26483478 24845553 30249741 21347285 20075255 24508170 28648823 29403074 27289101 20566863
18362917 19820115 25244985 26108605 25830018 25348373 24438588 22936249 15632085 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 11900973 23537049 21282665 17550304 20798317 26483478 24845553 30249741 21347285 20075255 24508170 28648823 29403074 27289101 20566863
EMBL
NWSH01002894
PCG67358.1
AGBW02009068
OWR51754.1
KQ459583
KPI98639.1
+ More
AGBW02014910 OWR40894.1 CH933808 EDW09756.2 AAAB01008966 EAA12953.3 AXCN02002114 CH916367 EDW01960.1 CH940648 EDW61204.1 KQ971363 KYB25710.1 JRES01000335 KNC32203.1 GBXI01007537 JAD06755.1 KYB25709.1 GAKP01003911 JAC55041.1 GDHF01031830 JAI20484.1 JXJN01019947 CH963849 EDW74481.1 ATLV01022351 KE525331 KFB47548.1 UFQS01001980 UFQT01001980 SSX12843.1 SSX32285.1 APCN01001942 APCN01001943 LBMM01001149 KMQ96820.1 CH479222 EDW34918.1 GBYB01009348 JAG79115.1 CM000363 CM002912 EDX10638.1 KMY99930.1 CH475417 EAL29340.3 GEZM01080345 JAV62208.1 CCAG010020383 OUUW01000029 SPP89779.1 AE014296 AGB94614.1 AY071594 NEVH01016943 PNF25141.1 KB632061 ERL88431.1 APGK01036285 KB740939 ENN77704.1 GL764503 EFZ17566.1 CM000159 EDW94958.1 CH954178 EDV52198.1 GL442829 EFN62856.1 KQ434796 KZC05639.1 JXUM01069301 KQ562544 KXJ75656.1 GEDC01028599 JAS08699.1 KK852871 KDR14580.1 KQ414615 KOC68651.1 QOIP01000001 RLU27277.1 ADTU01007672 ADTU01007673 ADTU01007674 ADTU01007675 GFXV01000576 MBW12381.1 KQ764883 OAD54273.1 KQ435711 KOX79401.1 KK107087 EZA59960.1 KQ976467 KYM84098.1 KZ288340 PBC27737.1 KQ981905 KYN33500.1 NNAY01003939 OXU18624.1 KQ977935 KYM98620.1 PYGN01000333 PSN48351.1 GGMS01003967 MBY73170.1 KQ980713 KYN14343.1 KQ982649 KYQ53019.1 CVRI01000043 CRK95867.1 ABLF02025046 GGMS01000184 MBY69387.1 GDRN01074479 GDRN01074475 JAI63234.1 GDRN01074476 JAI63236.1 GDRN01074477 JAI63235.1 GDRN01074473 JAI63238.1 IACF01003482 LAB69098.1 LJIJ01000043 ODN04440.1 DS235088 EEB11553.1
AGBW02014910 OWR40894.1 CH933808 EDW09756.2 AAAB01008966 EAA12953.3 AXCN02002114 CH916367 EDW01960.1 CH940648 EDW61204.1 KQ971363 KYB25710.1 JRES01000335 KNC32203.1 GBXI01007537 JAD06755.1 KYB25709.1 GAKP01003911 JAC55041.1 GDHF01031830 JAI20484.1 JXJN01019947 CH963849 EDW74481.1 ATLV01022351 KE525331 KFB47548.1 UFQS01001980 UFQT01001980 SSX12843.1 SSX32285.1 APCN01001942 APCN01001943 LBMM01001149 KMQ96820.1 CH479222 EDW34918.1 GBYB01009348 JAG79115.1 CM000363 CM002912 EDX10638.1 KMY99930.1 CH475417 EAL29340.3 GEZM01080345 JAV62208.1 CCAG010020383 OUUW01000029 SPP89779.1 AE014296 AGB94614.1 AY071594 NEVH01016943 PNF25141.1 KB632061 ERL88431.1 APGK01036285 KB740939 ENN77704.1 GL764503 EFZ17566.1 CM000159 EDW94958.1 CH954178 EDV52198.1 GL442829 EFN62856.1 KQ434796 KZC05639.1 JXUM01069301 KQ562544 KXJ75656.1 GEDC01028599 JAS08699.1 KK852871 KDR14580.1 KQ414615 KOC68651.1 QOIP01000001 RLU27277.1 ADTU01007672 ADTU01007673 ADTU01007674 ADTU01007675 GFXV01000576 MBW12381.1 KQ764883 OAD54273.1 KQ435711 KOX79401.1 KK107087 EZA59960.1 KQ976467 KYM84098.1 KZ288340 PBC27737.1 KQ981905 KYN33500.1 NNAY01003939 OXU18624.1 KQ977935 KYM98620.1 PYGN01000333 PSN48351.1 GGMS01003967 MBY73170.1 KQ980713 KYN14343.1 KQ982649 KYQ53019.1 CVRI01000043 CRK95867.1 ABLF02025046 GGMS01000184 MBY69387.1 GDRN01074479 GDRN01074475 JAI63234.1 GDRN01074476 JAI63236.1 GDRN01074477 JAI63235.1 GDRN01074473 JAI63238.1 IACF01003482 LAB69098.1 LJIJ01000043 ODN04440.1 DS235088 EEB11553.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000009192
UP000007062
UP000075903
+ More
UP000075902 UP000075886 UP000095301 UP000075882 UP000001070 UP000008792 UP000095300 UP000076407 UP000075880 UP000007266 UP000076408 UP000037069 UP000092460 UP000007798 UP000075885 UP000030765 UP000075920 UP000069272 UP000075840 UP000036403 UP000075900 UP000008744 UP000000304 UP000001819 UP000092445 UP000092444 UP000268350 UP000000803 UP000235965 UP000192223 UP000030742 UP000019118 UP000002282 UP000192221 UP000008711 UP000000311 UP000076502 UP000069940 UP000249989 UP000027135 UP000053825 UP000279307 UP000005205 UP000002358 UP000053105 UP000053097 UP000078540 UP000242457 UP000078541 UP000215335 UP000078542 UP000245037 UP000078492 UP000078200 UP000075809 UP000183832 UP000075881 UP000007819 UP000094527 UP000009046
UP000075902 UP000075886 UP000095301 UP000075882 UP000001070 UP000008792 UP000095300 UP000076407 UP000075880 UP000007266 UP000076408 UP000037069 UP000092460 UP000007798 UP000075885 UP000030765 UP000075920 UP000069272 UP000075840 UP000036403 UP000075900 UP000008744 UP000000304 UP000001819 UP000092445 UP000092444 UP000268350 UP000000803 UP000235965 UP000192223 UP000030742 UP000019118 UP000002282 UP000192221 UP000008711 UP000000311 UP000076502 UP000069940 UP000249989 UP000027135 UP000053825 UP000279307 UP000005205 UP000002358 UP000053105 UP000053097 UP000078540 UP000242457 UP000078541 UP000215335 UP000078542 UP000245037 UP000078492 UP000078200 UP000075809 UP000183832 UP000075881 UP000007819 UP000094527 UP000009046
PRIDE
Interpro
IPR005018
DOMON_domain
+ More
IPR014784 Cu2_ascorb_mOase-like_C
IPR036939 Cu2_ascorb_mOase_N_sf
IPR028460 Tbh/DBH
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR000323 Cu2_ascorb_mOase_N
IPR020611 Cu2_ascorb_mOase_CS-1
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR038751 Int8
IPR038697 CHD_cytochrome_sf
IPR014784 Cu2_ascorb_mOase-like_C
IPR036939 Cu2_ascorb_mOase_N_sf
IPR028460 Tbh/DBH
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR000323 Cu2_ascorb_mOase_N
IPR020611 Cu2_ascorb_mOase_CS-1
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR038751 Int8
IPR038697 CHD_cytochrome_sf
Gene 3D
ProteinModelPortal
A0A2A4J6Q1
A0A212FDE6
A0A194Q0W7
A0A212EHD6
B4KLU6
Q7Q325
+ More
A0A182VED0 A0A182U400 A0A182QCH8 A0A1I8M0I8 A0A182LPJ1 B4J6F9 A0A1I8M0I2 B4LPE5 A0A1I8PU30 A0A182XCB0 A0A182J9H7 A0A139WCR2 A0A182YDD4 A0A0L0CIG9 A0A0A1X7P4 A0A139WCN5 A0A034WHH5 A0A0K8U160 A0A1B0BT02 B4MQU2 A0A182P9D6 A0A084WBF4 A0A182WF50 A0A182FTC9 A0A336L841 A0A182HNL2 A0A0J7NWM9 A0A182RFB3 B4H8A3 A0A0C9Q5J7 B4QLT9 Q29CS1 A0A1A9ZYJ1 A0A1Y1KNQ2 A0A1B0G0W8 A0A3B0KSR7 M9PFT8 Q9VUY0 A0A2J7Q985 A0A1W4XC79 U4UCX0 N6TBP6 E9IPD3 B4PD26 A0A1W4W1E6 B3NDR5 E2AUK9 A0A154P125 A0A182GK54 A0A1B6C5X6 A0A067QW71 A0A0L7RD13 A0A3L8E3L6 A0A158P319 K7IPK7 A0A2H8TE63 A0A310SGK6 A0A0N0U712 A0A026WV84 A0A195BIF4 A0A2A3E7L9 A0A195EYX4 A0A232EJS7 A0A195CD26 A0A2P8YVT5 A0A2S2Q6E1 A0A195DN93 A0A1A9V9R5 A0A151WYR9 A0A1J1I6P5 A0A182K240 J9K956 A0A2S2PV31 A0A0P4W3V5 A0A0P4WP84 A0A0P4WAE3 A0A0P4WAC2 A0A2P2I4Z1 A0A1D2NGQ1 E0VDU7
A0A182VED0 A0A182U400 A0A182QCH8 A0A1I8M0I8 A0A182LPJ1 B4J6F9 A0A1I8M0I2 B4LPE5 A0A1I8PU30 A0A182XCB0 A0A182J9H7 A0A139WCR2 A0A182YDD4 A0A0L0CIG9 A0A0A1X7P4 A0A139WCN5 A0A034WHH5 A0A0K8U160 A0A1B0BT02 B4MQU2 A0A182P9D6 A0A084WBF4 A0A182WF50 A0A182FTC9 A0A336L841 A0A182HNL2 A0A0J7NWM9 A0A182RFB3 B4H8A3 A0A0C9Q5J7 B4QLT9 Q29CS1 A0A1A9ZYJ1 A0A1Y1KNQ2 A0A1B0G0W8 A0A3B0KSR7 M9PFT8 Q9VUY0 A0A2J7Q985 A0A1W4XC79 U4UCX0 N6TBP6 E9IPD3 B4PD26 A0A1W4W1E6 B3NDR5 E2AUK9 A0A154P125 A0A182GK54 A0A1B6C5X6 A0A067QW71 A0A0L7RD13 A0A3L8E3L6 A0A158P319 K7IPK7 A0A2H8TE63 A0A310SGK6 A0A0N0U712 A0A026WV84 A0A195BIF4 A0A2A3E7L9 A0A195EYX4 A0A232EJS7 A0A195CD26 A0A2P8YVT5 A0A2S2Q6E1 A0A195DN93 A0A1A9V9R5 A0A151WYR9 A0A1J1I6P5 A0A182K240 J9K956 A0A2S2PV31 A0A0P4W3V5 A0A0P4WP84 A0A0P4WAE3 A0A0P4WAC2 A0A2P2I4Z1 A0A1D2NGQ1 E0VDU7
PDB
4ZEL
E-value=7.51588e-52,
Score=518
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
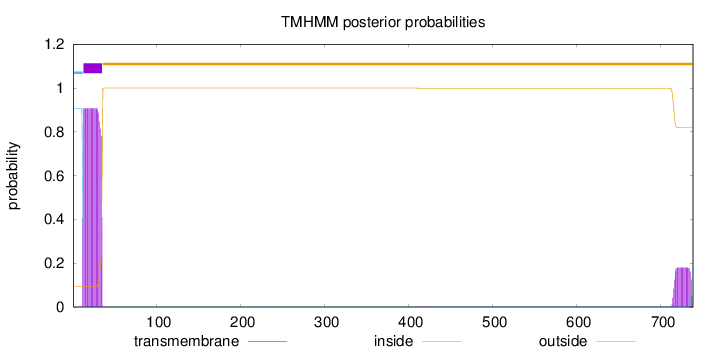
Length:
738
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.31315
Exp number, first 60 AAs:
20.47659
Total prob of N-in:
0.90747
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 738
Population Genetic Test Statistics
Pi
192.139483
Theta
161.758543
Tajima's D
1.015764
CLR
0.276011
CSRT
0.661766911654417
Interpretation
Uncertain
