Gene
KWMTBOMO01209
Annotation
antennal_esterase_CXE19_[Spodoptera_littoralis]
Full name
Carboxylic ester hydrolase
Location in the cell
Extracellular Reliability : 1.026 Nuclear Reliability : 1.212
Sequence
CDS
ATGTTGAACATAATAGTGTTGTTATTGCAAACGACTTTTATACTGGCAAGGGAGCCGAAACAAGTGACTTTGGTCAATCAAGGTACAATATCTGGAATGTACATTACGAGGTTTCGTACTAAACGTGTGGCCGCTTACGTTGGCGTTCCGTATGCTCAACCGCCGGTTGAATTTCGAAGATTTGCTGCTCCGGAAATCGCGAATCTGCCACAGTGGGAGGGCGTCAGGAATGCAACAATATTCGCGCCGGATTGCATGCAGAGGGTTCTTAAGAAAGAAGACTTGCAGAATCCATTGAAGAAGCACGATGAATTGTTTATGAAGCTACTAGAATCGCAGATGGAGGAGCCTAGGGAGAAAGAGTATTCGGAAGACTGTTTATACCTCAATGTCTACGTGCCTGATGGTGAGTAA
Protein
MLNIIVLLLQTTFILAREPKQVTLVNQGTISGMYITRFRTKRVAAYVGVPYAQPPVEFRRFAAPEIANLPQWEGVRNATIFAPDCMQRVLKKEDLQNPLKKHDELFMKLLESQMEEPREKEYSEDCLYLNVYVPDGE
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
A0A2H1WUL0
D3GDN0
A0A194PZ91
A0A2W1BPV5
A0A0N1II63
A0A2U3T8K7
+ More
A0A212FDD5 A0A182J3E2 A0A182S5T8 A0A182QQV5 A0A336LS68 A0A182M454 A0A084WEZ8 W5JTM4 A0A182U5M2 A0A182L3M4 A0A182RJM5 A0A182Y2D2 Q7QIM8 A0A182UWK1 A0A1J1HKP0 A0A1B0D5W3 A0A154P803 T1GJD5 A0A310SE26 D6W6X8 E2ACX6 A0A232EQY3 K7IYK8 A0A087ZZB6 E2BY80 A0A0L7R377 A0A026VUG0 A0A2A3EBX2 A0A2J7R4P4 A0A0C9RB64 A0A0M8ZRR2 A0A067R506 A0A151X477 N6TQC1 U4UX57 Q4V510 Q9W1B3 A0A0J9RLC1 A0A195CQU0 A0A195E0T4 A0A1W4UM93 B3NQ76 A0A1B0FJV8 E0VCL7 B4PAT9 A0A1B6I855 B4GCR3 Q28ZZ1 A0A1A9UJ11 A0A3B0IYX6 B3MDW8 A0A1B0A987 A0A0A1XM47 A0A1A9WP68 F4X3L5 A0A0L0C9E3 A0A195F311 B4KLV8 A0A195BLK0 A0A1B0BI94 A0A1A9Y6I7 A0A158P004 A0A2S2QQ08 A0A2R2MLM4 B4J6E6 A0A074ZLQ9 B4MQQ6 T1I8A1 A0A1S8WZ38 M3W106 G7YKI1 A0A2H1CRB2 A0A182YQK8 L8IPE1 A0A2J7QKJ9 B4MW86 A0A182R531 A0A2K5PKX1 A0A061I7X9 S9XI08 A0A061IAA7 A0A2J7QKK3 D2I5P4 A0A061I6Q8 A0A182VWB5 G1L8N4 A0A1U7TDB0 A0A0A7RQ48 A0A384CDR0 L9K1W8 A0A2J7QKK0 A0A2D0R6E4
A0A212FDD5 A0A182J3E2 A0A182S5T8 A0A182QQV5 A0A336LS68 A0A182M454 A0A084WEZ8 W5JTM4 A0A182U5M2 A0A182L3M4 A0A182RJM5 A0A182Y2D2 Q7QIM8 A0A182UWK1 A0A1J1HKP0 A0A1B0D5W3 A0A154P803 T1GJD5 A0A310SE26 D6W6X8 E2ACX6 A0A232EQY3 K7IYK8 A0A087ZZB6 E2BY80 A0A0L7R377 A0A026VUG0 A0A2A3EBX2 A0A2J7R4P4 A0A0C9RB64 A0A0M8ZRR2 A0A067R506 A0A151X477 N6TQC1 U4UX57 Q4V510 Q9W1B3 A0A0J9RLC1 A0A195CQU0 A0A195E0T4 A0A1W4UM93 B3NQ76 A0A1B0FJV8 E0VCL7 B4PAT9 A0A1B6I855 B4GCR3 Q28ZZ1 A0A1A9UJ11 A0A3B0IYX6 B3MDW8 A0A1B0A987 A0A0A1XM47 A0A1A9WP68 F4X3L5 A0A0L0C9E3 A0A195F311 B4KLV8 A0A195BLK0 A0A1B0BI94 A0A1A9Y6I7 A0A158P004 A0A2S2QQ08 A0A2R2MLM4 B4J6E6 A0A074ZLQ9 B4MQQ6 T1I8A1 A0A1S8WZ38 M3W106 G7YKI1 A0A2H1CRB2 A0A182YQK8 L8IPE1 A0A2J7QKJ9 B4MW86 A0A182R531 A0A2K5PKX1 A0A061I7X9 S9XI08 A0A061IAA7 A0A2J7QKK3 D2I5P4 A0A061I6Q8 A0A182VWB5 G1L8N4 A0A1U7TDB0 A0A0A7RQ48 A0A384CDR0 L9K1W8 A0A2J7QKK0 A0A2D0R6E4
EC Number
3.1.1.-
Pubmed
26354079
28756777
22118469
24438588
20920257
23761445
+ More
20966253 25244985 12364791 14747013 17210077 18362917 19820115 20798317 28648823 20075255 24508170 30249741 24845553 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17994087 20566863 17550304 15632085 25830018 21719571 26108605 21347285 17975172 22023798 22751099 23929341 23149746 20010809 24759410 23385571
20966253 25244985 12364791 14747013 17210077 18362917 19820115 20798317 28648823 20075255 24508170 30249741 24845553 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17994087 20566863 17550304 15632085 25830018 21719571 26108605 21347285 17975172 22023798 22751099 23929341 23149746 20010809 24759410 23385571
EMBL
ODYU01011171
SOQ56748.1
FJ652462
ACV60246.1
KQ459583
KPI98641.1
+ More
KZ150020 PZC74906.1 KQ460366 KPJ15633.1 KX015861 ARM65390.1 AGBW02009068 OWR51756.1 AXCN02000558 UFQT01000063 SSX19277.1 AXCM01007682 ATLV01023268 KE525341 KFB48792.1 ADMH02000528 ETN66099.1 AAAB01008807 EAA04720.3 CVRI01000003 CRK87030.1 AJVK01000413 AJVK01000414 AJVK01000415 KQ434839 KZC07983.1 CAQQ02187025 KQ760801 OAD58990.1 KQ971307 EFA11441.1 GL438602 EFN68706.1 NNAY01002687 OXU20773.1 GL451420 EFN79359.1 KQ414663 KOC65279.1 KK107929 QOIP01000003 EZA47111.1 RLU24612.1 KZ288295 PBC28974.1 NEVH01007404 PNF35795.1 GBYB01005565 JAG75332.1 KQ435885 KOX69617.1 KK852888 KDR14322.1 KQ982557 KYQ55084.1 APGK01024773 KB740562 ENN80248.1 KB632399 ERL94845.1 BT022846 AAY55262.1 AE013599 AAF47159.1 CM002911 KMY96254.1 KQ977394 KYN03010.1 KQ979955 KYN18542.1 CH954179 EDV56949.1 CCAG010018850 DS235059 EEB11123.1 CM000158 EDW92479.1 GECU01024585 JAS83121.1 CH479181 EDW32476.1 CM000071 EAL25471.1 OUUW01000001 SPP73235.1 CH902619 EDV37513.1 GBXI01002639 JAD11653.1 GL888624 EGI58812.1 JRES01000737 KNC28855.1 KQ981856 KYN34567.1 CH933808 EDW09768.2 KQ976440 KYM86578.1 JXJN01014867 ADTU01005183 ADTU01005184 GGMS01010591 MBY79794.1 CH916367 EDW01947.1 KL596709 KER28016.1 CH963849 EDW74445.1 ACPB03023208 KV893128 OON19671.1 AANG04002512 DF143501 GAA53463.1 KZ428635 PIS89960.1 JH881043 ELR57012.1 NEVH01013262 PNF29115.1 CH963857 EDW75956.2 KE673157 ERE78152.1 KB017973 KB017109 EPY74806.1 EPY80101.1 ERE78153.1 PNF29116.1 GL194740 EFB22645.1 ERE78151.1 ACTA01020472 KP141775 AJA38018.1 KB320919 ELW56780.1 PNF29114.1
KZ150020 PZC74906.1 KQ460366 KPJ15633.1 KX015861 ARM65390.1 AGBW02009068 OWR51756.1 AXCN02000558 UFQT01000063 SSX19277.1 AXCM01007682 ATLV01023268 KE525341 KFB48792.1 ADMH02000528 ETN66099.1 AAAB01008807 EAA04720.3 CVRI01000003 CRK87030.1 AJVK01000413 AJVK01000414 AJVK01000415 KQ434839 KZC07983.1 CAQQ02187025 KQ760801 OAD58990.1 KQ971307 EFA11441.1 GL438602 EFN68706.1 NNAY01002687 OXU20773.1 GL451420 EFN79359.1 KQ414663 KOC65279.1 KK107929 QOIP01000003 EZA47111.1 RLU24612.1 KZ288295 PBC28974.1 NEVH01007404 PNF35795.1 GBYB01005565 JAG75332.1 KQ435885 KOX69617.1 KK852888 KDR14322.1 KQ982557 KYQ55084.1 APGK01024773 KB740562 ENN80248.1 KB632399 ERL94845.1 BT022846 AAY55262.1 AE013599 AAF47159.1 CM002911 KMY96254.1 KQ977394 KYN03010.1 KQ979955 KYN18542.1 CH954179 EDV56949.1 CCAG010018850 DS235059 EEB11123.1 CM000158 EDW92479.1 GECU01024585 JAS83121.1 CH479181 EDW32476.1 CM000071 EAL25471.1 OUUW01000001 SPP73235.1 CH902619 EDV37513.1 GBXI01002639 JAD11653.1 GL888624 EGI58812.1 JRES01000737 KNC28855.1 KQ981856 KYN34567.1 CH933808 EDW09768.2 KQ976440 KYM86578.1 JXJN01014867 ADTU01005183 ADTU01005184 GGMS01010591 MBY79794.1 CH916367 EDW01947.1 KL596709 KER28016.1 CH963849 EDW74445.1 ACPB03023208 KV893128 OON19671.1 AANG04002512 DF143501 GAA53463.1 KZ428635 PIS89960.1 JH881043 ELR57012.1 NEVH01013262 PNF29115.1 CH963857 EDW75956.2 KE673157 ERE78152.1 KB017973 KB017109 EPY74806.1 EPY80101.1 ERE78153.1 PNF29116.1 GL194740 EFB22645.1 ERE78151.1 ACTA01020472 KP141775 AJA38018.1 KB320919 ELW56780.1 PNF29114.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000075880
UP000075901
UP000075886
+ More
UP000075883 UP000030765 UP000000673 UP000075902 UP000075882 UP000075900 UP000076408 UP000007062 UP000075903 UP000183832 UP000092462 UP000076502 UP000015102 UP000007266 UP000000311 UP000215335 UP000002358 UP000005203 UP000008237 UP000053825 UP000053097 UP000279307 UP000242457 UP000235965 UP000053105 UP000027135 UP000075809 UP000019118 UP000030742 UP000000803 UP000078542 UP000078492 UP000192221 UP000008711 UP000092444 UP000009046 UP000002282 UP000008744 UP000001819 UP000078200 UP000268350 UP000007801 UP000092445 UP000091820 UP000007755 UP000037069 UP000078541 UP000009192 UP000078540 UP000092460 UP000092443 UP000005205 UP000085678 UP000001070 UP000007798 UP000015103 UP000011712 UP000233040 UP000030759 UP000075920 UP000008912 UP000189704 UP000261680 UP000011518 UP000221080
UP000075883 UP000030765 UP000000673 UP000075902 UP000075882 UP000075900 UP000076408 UP000007062 UP000075903 UP000183832 UP000092462 UP000076502 UP000015102 UP000007266 UP000000311 UP000215335 UP000002358 UP000005203 UP000008237 UP000053825 UP000053097 UP000279307 UP000242457 UP000235965 UP000053105 UP000027135 UP000075809 UP000019118 UP000030742 UP000000803 UP000078542 UP000078492 UP000192221 UP000008711 UP000092444 UP000009046 UP000002282 UP000008744 UP000001819 UP000078200 UP000268350 UP000007801 UP000092445 UP000091820 UP000007755 UP000037069 UP000078541 UP000009192 UP000078540 UP000092460 UP000092443 UP000005205 UP000085678 UP000001070 UP000007798 UP000015103 UP000011712 UP000233040 UP000030759 UP000075920 UP000008912 UP000189704 UP000261680 UP000011518 UP000221080
Interpro
IPR019819
Carboxylesterase_B_CS
+ More
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR000997 Cholinesterase
IPR002168 Lipase_GDXG_HIS_AS
IPR030025 NLGN4
IPR000832 GPCR_2_secretin-like
IPR009071 HMG_box_dom
IPR018359 Bromodomain_CS
IPR037382 Rsc/polybromo
IPR001025 BAH_dom
IPR017981 GPCR_2-like
IPR001487 Bromodomain
IPR027180 RSC1/2/4
IPR036427 Bromodomain-like_sf
IPR036910 HMG_box_dom_sf
IPR037968 PBRM1_BD5
IPR000460 Nlgn
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR000997 Cholinesterase
IPR002168 Lipase_GDXG_HIS_AS
IPR030025 NLGN4
IPR000832 GPCR_2_secretin-like
IPR009071 HMG_box_dom
IPR018359 Bromodomain_CS
IPR037382 Rsc/polybromo
IPR001025 BAH_dom
IPR017981 GPCR_2-like
IPR001487 Bromodomain
IPR027180 RSC1/2/4
IPR036427 Bromodomain-like_sf
IPR036910 HMG_box_dom_sf
IPR037968 PBRM1_BD5
IPR000460 Nlgn
Gene 3D
ProteinModelPortal
A0A2H1WUL0
D3GDN0
A0A194PZ91
A0A2W1BPV5
A0A0N1II63
A0A2U3T8K7
+ More
A0A212FDD5 A0A182J3E2 A0A182S5T8 A0A182QQV5 A0A336LS68 A0A182M454 A0A084WEZ8 W5JTM4 A0A182U5M2 A0A182L3M4 A0A182RJM5 A0A182Y2D2 Q7QIM8 A0A182UWK1 A0A1J1HKP0 A0A1B0D5W3 A0A154P803 T1GJD5 A0A310SE26 D6W6X8 E2ACX6 A0A232EQY3 K7IYK8 A0A087ZZB6 E2BY80 A0A0L7R377 A0A026VUG0 A0A2A3EBX2 A0A2J7R4P4 A0A0C9RB64 A0A0M8ZRR2 A0A067R506 A0A151X477 N6TQC1 U4UX57 Q4V510 Q9W1B3 A0A0J9RLC1 A0A195CQU0 A0A195E0T4 A0A1W4UM93 B3NQ76 A0A1B0FJV8 E0VCL7 B4PAT9 A0A1B6I855 B4GCR3 Q28ZZ1 A0A1A9UJ11 A0A3B0IYX6 B3MDW8 A0A1B0A987 A0A0A1XM47 A0A1A9WP68 F4X3L5 A0A0L0C9E3 A0A195F311 B4KLV8 A0A195BLK0 A0A1B0BI94 A0A1A9Y6I7 A0A158P004 A0A2S2QQ08 A0A2R2MLM4 B4J6E6 A0A074ZLQ9 B4MQQ6 T1I8A1 A0A1S8WZ38 M3W106 G7YKI1 A0A2H1CRB2 A0A182YQK8 L8IPE1 A0A2J7QKJ9 B4MW86 A0A182R531 A0A2K5PKX1 A0A061I7X9 S9XI08 A0A061IAA7 A0A2J7QKK3 D2I5P4 A0A061I6Q8 A0A182VWB5 G1L8N4 A0A1U7TDB0 A0A0A7RQ48 A0A384CDR0 L9K1W8 A0A2J7QKK0 A0A2D0R6E4
A0A212FDD5 A0A182J3E2 A0A182S5T8 A0A182QQV5 A0A336LS68 A0A182M454 A0A084WEZ8 W5JTM4 A0A182U5M2 A0A182L3M4 A0A182RJM5 A0A182Y2D2 Q7QIM8 A0A182UWK1 A0A1J1HKP0 A0A1B0D5W3 A0A154P803 T1GJD5 A0A310SE26 D6W6X8 E2ACX6 A0A232EQY3 K7IYK8 A0A087ZZB6 E2BY80 A0A0L7R377 A0A026VUG0 A0A2A3EBX2 A0A2J7R4P4 A0A0C9RB64 A0A0M8ZRR2 A0A067R506 A0A151X477 N6TQC1 U4UX57 Q4V510 Q9W1B3 A0A0J9RLC1 A0A195CQU0 A0A195E0T4 A0A1W4UM93 B3NQ76 A0A1B0FJV8 E0VCL7 B4PAT9 A0A1B6I855 B4GCR3 Q28ZZ1 A0A1A9UJ11 A0A3B0IYX6 B3MDW8 A0A1B0A987 A0A0A1XM47 A0A1A9WP68 F4X3L5 A0A0L0C9E3 A0A195F311 B4KLV8 A0A195BLK0 A0A1B0BI94 A0A1A9Y6I7 A0A158P004 A0A2S2QQ08 A0A2R2MLM4 B4J6E6 A0A074ZLQ9 B4MQQ6 T1I8A1 A0A1S8WZ38 M3W106 G7YKI1 A0A2H1CRB2 A0A182YQK8 L8IPE1 A0A2J7QKJ9 B4MW86 A0A182R531 A0A2K5PKX1 A0A061I7X9 S9XI08 A0A061IAA7 A0A2J7QKK3 D2I5P4 A0A061I6Q8 A0A182VWB5 G1L8N4 A0A1U7TDB0 A0A0A7RQ48 A0A384CDR0 L9K1W8 A0A2J7QKK0 A0A2D0R6E4
PDB
6EMI
E-value=2.5668e-13,
Score=176
Ontologies
KEGG
GO
GO:0050804
GO:0045202
GO:0098793
GO:0042043
GO:0005887
GO:0097105
GO:0009986
GO:0038023
GO:0097104
GO:0007158
GO:0048488
GO:0016787
GO:0016021
GO:0080030
GO:0004104
GO:0004771
GO:0016042
GO:0004806
GO:0005615
GO:0052689
GO:0007155
GO:0050808
GO:0003682
GO:0007166
GO:0006338
GO:0004930
GO:0003677
GO:0016586
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.806048
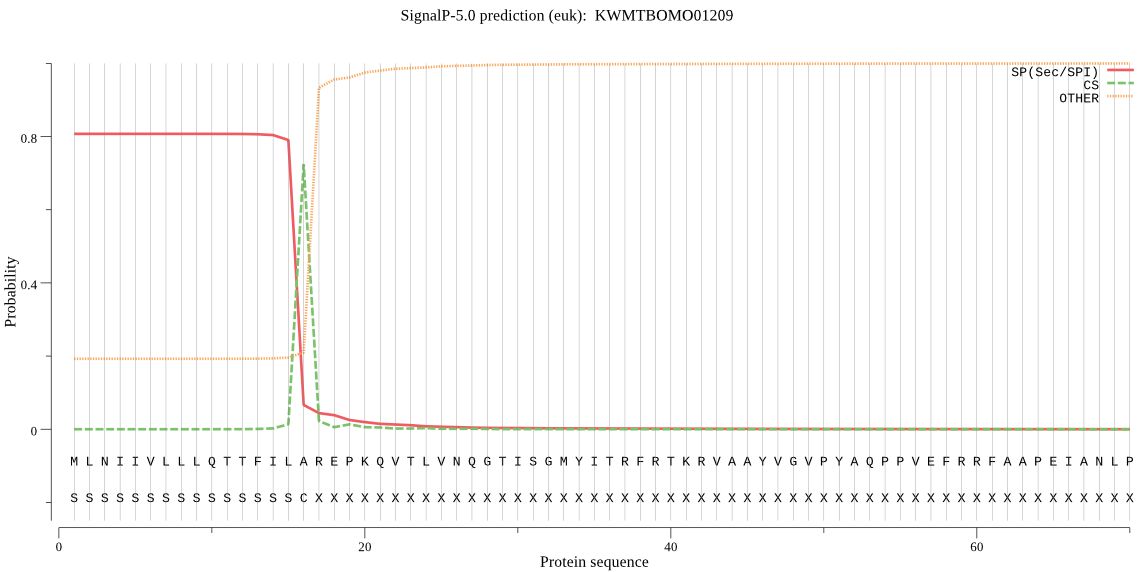
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00793
Exp number, first 60 AAs:
0.00787
Total prob of N-in:
0.02832
outside
1 - 137

Population Genetic Test Statistics
Pi
330.535637
Theta
207.621208
Tajima's D
1.863728
CLR
0.15714
CSRT
0.859907004649768
Interpretation
Uncertain
