Gene
KWMTBOMO01206
Pre Gene Modal
BGIBMGA008810
Annotation
PREDICTED:_uncharacterized_protein_LOC106143374_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.622 Nuclear Reliability : 1.043
Sequence
CDS
ATGAAGATTACAGTACTCTGCGTAGCTATGATGGTTATTTGCTGCTCGGCCCAACGTTCGCCATACGCTGGAAGACAACCAATAGGTTTTCCTGCAATTCAAAGCACGGCACCCCCAGAAGACGCTTTGGGCAATAGATTCGGAGACGATATCAGTACGACGACAATACGACTCCCGATCGAAGCTTTGGGTGACGCTGACCTGGTGAATCGACTAAGAAAACTGCCGGTCGACAAGCAGCCGTTTTGGCTCCTCAACTGGCTGGCTTTGGAAGCCAACAGGAAGAATCCACAGAATTTCAATCAAAGACCGAATAGTTTTCTCGATAACTCCATTAGGAATGTTTAA
Protein
MKITVLCVAMMVICCSAQRSPYAGRQPIGFPAIQSTAPPEDALGNRFGDDISTTTIRLPIEALGDADLVNRLRKLPVDKQPFWLLNWLALEANRKNPQNFNQRPNSFLDNSIRNV
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JH13
A0A2H1W2U5
A0A2W1BT59
A0A2A4ITL1
A0A212FD86
I4DKI1
+ More
H9JY46 A0A2A4J647 A0A2W1BMR5 S4Q037 A0A194PZ81 A0A212FDC0 H9JH06 A0A0N1II63 A0A2H1VLA6 A0A194Q025 A0A2A4J5R5 A0A2H1WBD2 A0A212FD96 A0A2W1BKV4 A0A212FD67 A0A0L7LGY0 A0A0N1IGY2 D9HQ52 A0A212FD84 A0A1E1W4D1 A0A1J1HJY7 A0A212FD81 I4DJU7 H9JH12 A0A0T6AVL6 A0A194QIR8 A0A0N0PD66 D6W7B5 Q7KTW1 A0A2W1BMS0 A0A2H1VV24 A0A195BVN9 A0A158N937 B4QK62 A0A151X4M8 A0A1W4WQ17 A0A194Q0X2 A0A151J817 A0A0N0BFR9 B4IAR8 B3NEA8 N6UQQ5 A0A1B0BPC2 A0A1A9XDB3 A0A1B0FFC0 A0A1A9ZS57 A0A1B6CJE4 A0A088ALH7 T1HZ27 B4ITD5 A0A1Y1K4Z1 A0A1A9UQJ5 V9IJ57 B4PD42 A0A2A3E8I5 B3NIZ0 A0A1Y1LT35 A0A2K8JRM1 A0A2P8ZIL0 A0A0K8US98 A0A0L7L5R2 A0A2J7R6Y7 A0A0A1WZ51
H9JY46 A0A2A4J647 A0A2W1BMR5 S4Q037 A0A194PZ81 A0A212FDC0 H9JH06 A0A0N1II63 A0A2H1VLA6 A0A194Q025 A0A2A4J5R5 A0A2H1WBD2 A0A212FD96 A0A2W1BKV4 A0A212FD67 A0A0L7LGY0 A0A0N1IGY2 D9HQ52 A0A212FD84 A0A1E1W4D1 A0A1J1HJY7 A0A212FD81 I4DJU7 H9JH12 A0A0T6AVL6 A0A194QIR8 A0A0N0PD66 D6W7B5 Q7KTW1 A0A2W1BMS0 A0A2H1VV24 A0A195BVN9 A0A158N937 B4QK62 A0A151X4M8 A0A1W4WQ17 A0A194Q0X2 A0A151J817 A0A0N0BFR9 B4IAR8 B3NEA8 N6UQQ5 A0A1B0BPC2 A0A1A9XDB3 A0A1B0FFC0 A0A1A9ZS57 A0A1B6CJE4 A0A088ALH7 T1HZ27 B4ITD5 A0A1Y1K4Z1 A0A1A9UQJ5 V9IJ57 B4PD42 A0A2A3E8I5 B3NIZ0 A0A1Y1LT35 A0A2K8JRM1 A0A2P8ZIL0 A0A0K8US98 A0A0L7L5R2 A0A2J7R6Y7 A0A0A1WZ51
Pubmed
EMBL
BABH01039328
ODYU01005969
SOQ47409.1
KZ150020
PZC74913.1
NWSH01007163
+ More
PCG63061.1 AGBW02009098 OWR51690.1 AK401799 KQ459583 BAM18421.1 KPI98640.1 BABH01042699 NWSH01002894 PCG67359.1 PZC74915.1 GAIX01000468 JAA92092.1 KPI98642.1 AGBW02009068 OWR51755.1 BABH01039338 KQ460366 KPJ15633.1 ODYU01003176 SOQ41586.1 KPI98643.1 NWSH01002910 PCG67311.1 ODYU01007537 SOQ50395.1 OWR51693.1 PZC74911.1 OWR51695.1 JTDY01001174 KOB74650.1 KPJ15635.1 HM023807 ADJ58540.1 OWR51692.1 GDQN01009224 JAT81830.1 CVRI01000001 CRK86561.1 OWR51694.1 AK401565 BAM18187.1 LJIG01022763 KRT78846.1 KQ458860 KPJ04825.1 KPJ15631.1 KQ971307 EFA11052.1 AE014296 BT030906 BT030927 BK002263 AAS65091.1 ABV82288.1 ABV82309.1 DAA03106.1 PZC74914.1 ODYU01004608 SOQ44673.1 KQ976405 KYM91861.1 ADTU01009064 CM000363 CM002912 EDX11399.1 KMZ01040.1 KQ982543 KYQ55362.1 KPI98644.1 KQ979640 KYN20051.1 KQ435794 KOX73738.1 CH480826 EDW44381.1 CH954178 EDV52672.1 APGK01020323 KB740193 KB631669 ENN81062.1 ERL85175.1 JXJN01017882 CCAG010000766 GEDC01023694 JAS13604.1 ACPB03014189 CH891690 EDW99648.1 GEZM01095099 JAV55508.1 JR049756 AEY61065.1 CM000159 EDW94974.1 KZ288324 PBC28083.1 EDV52707.1 GEZM01047546 JAV76812.1 KY030929 ATU82680.1 PYGN01000046 PSN56310.1 GDHF01022863 JAI29451.1 JTDY01002715 KOB70863.1 NEVH01006740 PNF36593.1 GBXI01010599 JAD03693.1
PCG63061.1 AGBW02009098 OWR51690.1 AK401799 KQ459583 BAM18421.1 KPI98640.1 BABH01042699 NWSH01002894 PCG67359.1 PZC74915.1 GAIX01000468 JAA92092.1 KPI98642.1 AGBW02009068 OWR51755.1 BABH01039338 KQ460366 KPJ15633.1 ODYU01003176 SOQ41586.1 KPI98643.1 NWSH01002910 PCG67311.1 ODYU01007537 SOQ50395.1 OWR51693.1 PZC74911.1 OWR51695.1 JTDY01001174 KOB74650.1 KPJ15635.1 HM023807 ADJ58540.1 OWR51692.1 GDQN01009224 JAT81830.1 CVRI01000001 CRK86561.1 OWR51694.1 AK401565 BAM18187.1 LJIG01022763 KRT78846.1 KQ458860 KPJ04825.1 KPJ15631.1 KQ971307 EFA11052.1 AE014296 BT030906 BT030927 BK002263 AAS65091.1 ABV82288.1 ABV82309.1 DAA03106.1 PZC74914.1 ODYU01004608 SOQ44673.1 KQ976405 KYM91861.1 ADTU01009064 CM000363 CM002912 EDX11399.1 KMZ01040.1 KQ982543 KYQ55362.1 KPI98644.1 KQ979640 KYN20051.1 KQ435794 KOX73738.1 CH480826 EDW44381.1 CH954178 EDV52672.1 APGK01020323 KB740193 KB631669 ENN81062.1 ERL85175.1 JXJN01017882 CCAG010000766 GEDC01023694 JAS13604.1 ACPB03014189 CH891690 EDW99648.1 GEZM01095099 JAV55508.1 JR049756 AEY61065.1 CM000159 EDW94974.1 KZ288324 PBC28083.1 EDV52707.1 GEZM01047546 JAV76812.1 KY030929 ATU82680.1 PYGN01000046 PSN56310.1 GDHF01022863 JAI29451.1 JTDY01002715 KOB70863.1 NEVH01006740 PNF36593.1 GBXI01010599 JAD03693.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000183832 UP000007266 UP000000803 UP000078540 UP000005205 UP000000304 UP000075809 UP000192223 UP000078492 UP000053105 UP000001292 UP000008711 UP000019118 UP000030742 UP000092460 UP000092443 UP000092444 UP000092445 UP000005203 UP000015103 UP000002282 UP000078200 UP000242457 UP000245037 UP000235965
UP000183832 UP000007266 UP000000803 UP000078540 UP000005205 UP000000304 UP000075809 UP000192223 UP000078492 UP000053105 UP000001292 UP000008711 UP000019118 UP000030742 UP000092460 UP000092443 UP000092444 UP000092445 UP000005203 UP000015103 UP000002282 UP000078200 UP000242457 UP000245037 UP000235965
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JH13
A0A2H1W2U5
A0A2W1BT59
A0A2A4ITL1
A0A212FD86
I4DKI1
+ More
H9JY46 A0A2A4J647 A0A2W1BMR5 S4Q037 A0A194PZ81 A0A212FDC0 H9JH06 A0A0N1II63 A0A2H1VLA6 A0A194Q025 A0A2A4J5R5 A0A2H1WBD2 A0A212FD96 A0A2W1BKV4 A0A212FD67 A0A0L7LGY0 A0A0N1IGY2 D9HQ52 A0A212FD84 A0A1E1W4D1 A0A1J1HJY7 A0A212FD81 I4DJU7 H9JH12 A0A0T6AVL6 A0A194QIR8 A0A0N0PD66 D6W7B5 Q7KTW1 A0A2W1BMS0 A0A2H1VV24 A0A195BVN9 A0A158N937 B4QK62 A0A151X4M8 A0A1W4WQ17 A0A194Q0X2 A0A151J817 A0A0N0BFR9 B4IAR8 B3NEA8 N6UQQ5 A0A1B0BPC2 A0A1A9XDB3 A0A1B0FFC0 A0A1A9ZS57 A0A1B6CJE4 A0A088ALH7 T1HZ27 B4ITD5 A0A1Y1K4Z1 A0A1A9UQJ5 V9IJ57 B4PD42 A0A2A3E8I5 B3NIZ0 A0A1Y1LT35 A0A2K8JRM1 A0A2P8ZIL0 A0A0K8US98 A0A0L7L5R2 A0A2J7R6Y7 A0A0A1WZ51
H9JY46 A0A2A4J647 A0A2W1BMR5 S4Q037 A0A194PZ81 A0A212FDC0 H9JH06 A0A0N1II63 A0A2H1VLA6 A0A194Q025 A0A2A4J5R5 A0A2H1WBD2 A0A212FD96 A0A2W1BKV4 A0A212FD67 A0A0L7LGY0 A0A0N1IGY2 D9HQ52 A0A212FD84 A0A1E1W4D1 A0A1J1HJY7 A0A212FD81 I4DJU7 H9JH12 A0A0T6AVL6 A0A194QIR8 A0A0N0PD66 D6W7B5 Q7KTW1 A0A2W1BMS0 A0A2H1VV24 A0A195BVN9 A0A158N937 B4QK62 A0A151X4M8 A0A1W4WQ17 A0A194Q0X2 A0A151J817 A0A0N0BFR9 B4IAR8 B3NEA8 N6UQQ5 A0A1B0BPC2 A0A1A9XDB3 A0A1B0FFC0 A0A1A9ZS57 A0A1B6CJE4 A0A088ALH7 T1HZ27 B4ITD5 A0A1Y1K4Z1 A0A1A9UQJ5 V9IJ57 B4PD42 A0A2A3E8I5 B3NIZ0 A0A1Y1LT35 A0A2K8JRM1 A0A2P8ZIL0 A0A0K8US98 A0A0L7L5R2 A0A2J7R6Y7 A0A0A1WZ51
Ontologies
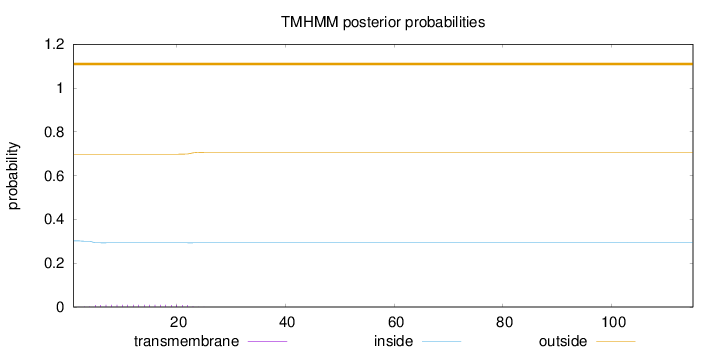
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.994035
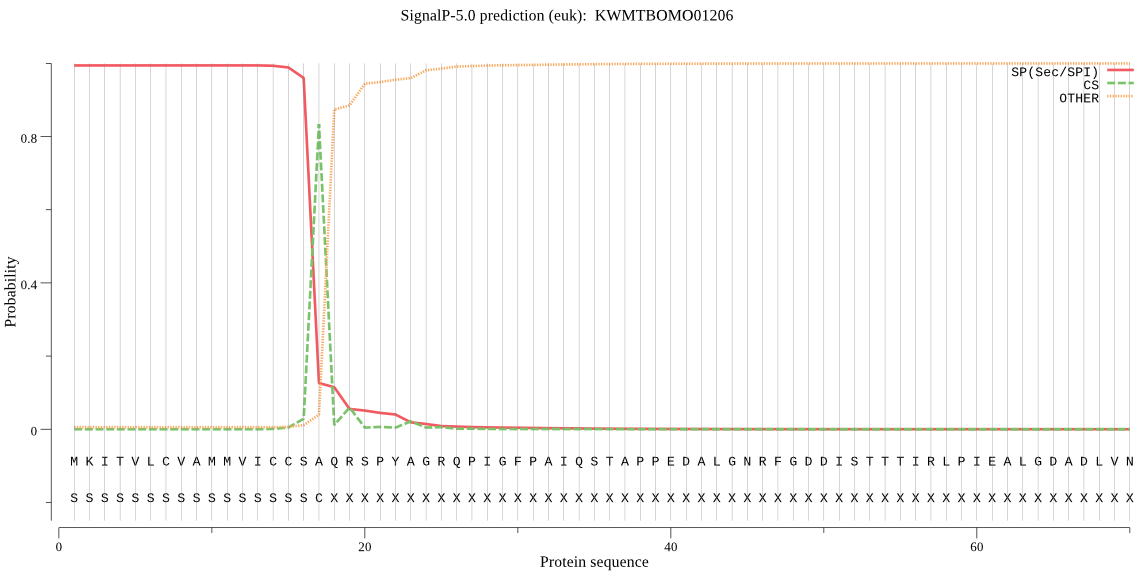
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17318
Exp number, first 60 AAs:
0.17318
Total prob of N-in:
0.30211
outside
1 - 115
Population Genetic Test Statistics
Pi
237.424459
Theta
159.837815
Tajima's D
1.496917
CLR
0
CSRT
0.784610769461527
Interpretation
Uncertain
