Gene
KWMTBOMO01196
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.659
Sequence
CDS
ATGCTGGGAGTATCCAACAGATCGAAGCGGTACTTCACGCCTGGACAAAGGCTTTTGCTTTATAAAGCACAAGTCCGGCCTCGAGTGGAGTACTGCTCCCATCTCTGGGCTGGGGCTCCCAAATACCAGCTTCTTCCATTGGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATTATCCCATTCTCACGGATCGTTTGGAACCTCTGGGTCTGCGGAGGGACTTCGGTTCCCTCTGTGTTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCGGCATCTCGTTTTTACCATCGCACCGCCCGCCACCGGAGTAGAGCTCATCCATACTACATGGAGCCACTGCGGTCATCCACAGTGCGTTTCCAGAGGTATTTTTGCCACGTACCATCCGGCTAA
Protein
MLGVSNRSKRYFTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPLDSIQRRAVRIVDYPILTDRLEPLGLRRDFGSLCVLYRMFHGECSEELFEMIPASRFYHRTARHRSRAHPYYMEPLRSSTVRFQRYFCHVPSG
Summary
Uniprot
Q5KTM5
Q6UV17
A0A2H1WAR9
A0A2H1VFZ3
B9WPT5
T2M6M7
+ More
A0A2H1V2F4 J9GT67 A0A2H1WCG9 A0A3D5S1K1 A0A2H1VUV7 A0A0P4VXU0 A0A2H1WY65 T2MGM5 A0A1Y1S3P2 A0A2H1W7F5 A0A1E1XGQ7 A0A093I968 A0A093H5S5 A0A093JFI6 A0A093HMV3 A0A093GU34 A0A093IBP9 A0A093JL05 A0A093H7K6 A0A093HHB4 A0A093HVG8 A0A093JHV8 A0A093HF89 A0A093HFB7 A0A093HCT5 A0A093HJX0 A0A093HT80 A0A093HYW2 A0A093HBM3 A0A093JC36 A0A093HPP7 A0A093HNU6 A0A093HPY1 A0A093HPA2 A0A093I7Q5 A0A093GVA5 A0A093HMV9 A0A093J7J9 A0A093HGI5 A0A093JU31 A0A093HW56 A0A093HDI4 A0A093GW31 A0A093HRW3 A0A093GQT0 A0A093HB81 A0A093HVT9 A0A093JR35 A0A093H7L9 A0A093HWQ4 A0A3M0IVW1 A0A093HHZ8 A0A093HDI8 A0A3M0JRY7 A0A2G8JWN9 A0A093HQY8 A0A093IC93 A0A093HQN0 A0A093HRM3 A0A093HJW5 A0A3M0KD23 A0A093HP87 A0A3M0L9G9 A0A3M0JVZ5 A0A093JJ50 A0A093IA31 A0A3M0KPI8 A0A093H881 A0A093GWX3 A0A093HJY5 A0A093HWI0 A0A093K6G3 A0A093HPR5 A0A093H950 A0A3M0L7E2 A0A093HGX5 A0A093HAN0 A0A093JWX0 A0A093J469 A0A093H6R6 A0A091HUB5
A0A2H1V2F4 J9GT67 A0A2H1WCG9 A0A3D5S1K1 A0A2H1VUV7 A0A0P4VXU0 A0A2H1WY65 T2MGM5 A0A1Y1S3P2 A0A2H1W7F5 A0A1E1XGQ7 A0A093I968 A0A093H5S5 A0A093JFI6 A0A093HMV3 A0A093GU34 A0A093IBP9 A0A093JL05 A0A093H7K6 A0A093HHB4 A0A093HVG8 A0A093JHV8 A0A093HF89 A0A093HFB7 A0A093HCT5 A0A093HJX0 A0A093HT80 A0A093HYW2 A0A093HBM3 A0A093JC36 A0A093HPP7 A0A093HNU6 A0A093HPY1 A0A093HPA2 A0A093I7Q5 A0A093GVA5 A0A093HMV9 A0A093J7J9 A0A093HGI5 A0A093JU31 A0A093HW56 A0A093HDI4 A0A093GW31 A0A093HRW3 A0A093GQT0 A0A093HB81 A0A093HVT9 A0A093JR35 A0A093H7L9 A0A093HWQ4 A0A3M0IVW1 A0A093HHZ8 A0A093HDI8 A0A3M0JRY7 A0A2G8JWN9 A0A093HQY8 A0A093IC93 A0A093HQN0 A0A093HRM3 A0A093HJW5 A0A3M0KD23 A0A093HP87 A0A3M0L9G9 A0A3M0JVZ5 A0A093JJ50 A0A093IA31 A0A3M0KPI8 A0A093H881 A0A093GWX3 A0A093HJY5 A0A093HWI0 A0A093K6G3 A0A093HPR5 A0A093H950 A0A3M0L7E2 A0A093HGX5 A0A093HAN0 A0A093JWX0 A0A093J469 A0A093H6R6 A0A091HUB5
EMBL
AB126052
BAD86652.1
AY359886
AAQ57129.1
ODYU01007424
SOQ50185.1
+ More
ODYU01002162 SOQ39302.1 FM995623 CAX36785.1 HAAD01001474 CDG67706.1 ODYU01000366 SOQ35027.1 AMCI01002135 EJX03495.1 ODYU01007423 SOQ50184.1 DPOC01000269 HCX22453.1 ODYU01004409 SOQ44252.1 GDRN01105059 JAI57791.1 ODYU01011959 SOQ58001.1 HAAD01005022 CDG71254.1 LWDP01000391 ORD92879.1 ODYU01006795 SOQ48973.1 GFAC01000919 JAT98269.1 KL206974 KFV87692.1 KL206020 KFV77928.1 KL205964 KFV77614.1 KL206569 KFV83968.1 KL205741 KFV73823.1 KL206232 KL206309 KL206999 KFV80275.1 KFV81186.1 KFV88582.1 KL206163 KFV79444.1 KL205979 KFV77661.1 KL206307 KFV81126.1 KL206894 KFV86733.1 KL206048 KFV78374.1 KL206330 KFV81283.1 KL206333 KFV81308.1 KL206236 KFV80393.1 KL206502 KFV82928.1 KL206805 KFV85878.1 KL206613 KFV84187.1 KL206225 KFV80018.1 KL205903 KFV76469.1 KL206341 KFV81395.1 KL206264 KFV80687.1 KFV81495.1 KL206638 KFV84453.1 KL206936 KFV87212.1 KL205765 KFV74228.1 KL206526 KFV83081.1 KL205812 KFV74894.1 KL205728 KL205733 KL205757 KL205924 KL206049 KL206274 KL206359 KL206368 KL206416 KL206438 KL206458 KL206553 KL206738 KL206902 KL206932 KL206979 KL206995 KL206996 KL207002 KFV73492.1 KFV73612.1 KFV74069.1 KFV77013.1 KFV78397.1 KFV80821.1 KFV81678.1 KFV81842.1 KFV82263.1 KFV82322.1 KFV82483.1 KFV83356.1 KFV85496.1 KFV86827.1 KFV87138.1 KFV87825.1 KFV88414.1 KFV88462.1 KFV88705.1 KL206410 KFV82084.1 KL206549 KFV83237.1 KL205926 KFV77067.1 KFV73601.1 KL206733 KFV85403.1 KL205665 KFV72673.1 KL205888 KFV76262.1 KL206562 KFV83655.1 KFV81119.1 KL206071 KFV78608.1 KFV83965.1 QRBI01000220 RMB92612.1 KL206129 KFV79085.1 KL206190 KFV79766.1 QRBI01000131 RMC03475.1 MRZV01001153 PIK40187.1 KL206600 KFV84076.1 KL207003 KFV88757.1 KL206694 KFV84978.1 KL206630 KFV84296.1 KL206188 KFV79735.1 QRBI01000111 RMC10945.1 KL206273 KFV80807.1 QRBI01000093 RMC21953.1 QRBI01000126 RMC04361.1 KL206086 KFV78804.1 KFV88760.1 QRBI01000106 RMC12940.1 KL205829 KFV75232.1 KL205810 KFV74798.1 KL206513 KFV82948.1 KL206811 KFV86001.1 KL206812 KFV86049.1 KFV81420.1 KL205877 KFV75915.1 RMC21288.1 KL206301 KFV80966.1 KFV76460.1 KL206514 KFV82949.1 KL205740 KFV73754.1 KL206036 KFV78283.1 KL217824 KFO99526.1
ODYU01002162 SOQ39302.1 FM995623 CAX36785.1 HAAD01001474 CDG67706.1 ODYU01000366 SOQ35027.1 AMCI01002135 EJX03495.1 ODYU01007423 SOQ50184.1 DPOC01000269 HCX22453.1 ODYU01004409 SOQ44252.1 GDRN01105059 JAI57791.1 ODYU01011959 SOQ58001.1 HAAD01005022 CDG71254.1 LWDP01000391 ORD92879.1 ODYU01006795 SOQ48973.1 GFAC01000919 JAT98269.1 KL206974 KFV87692.1 KL206020 KFV77928.1 KL205964 KFV77614.1 KL206569 KFV83968.1 KL205741 KFV73823.1 KL206232 KL206309 KL206999 KFV80275.1 KFV81186.1 KFV88582.1 KL206163 KFV79444.1 KL205979 KFV77661.1 KL206307 KFV81126.1 KL206894 KFV86733.1 KL206048 KFV78374.1 KL206330 KFV81283.1 KL206333 KFV81308.1 KL206236 KFV80393.1 KL206502 KFV82928.1 KL206805 KFV85878.1 KL206613 KFV84187.1 KL206225 KFV80018.1 KL205903 KFV76469.1 KL206341 KFV81395.1 KL206264 KFV80687.1 KFV81495.1 KL206638 KFV84453.1 KL206936 KFV87212.1 KL205765 KFV74228.1 KL206526 KFV83081.1 KL205812 KFV74894.1 KL205728 KL205733 KL205757 KL205924 KL206049 KL206274 KL206359 KL206368 KL206416 KL206438 KL206458 KL206553 KL206738 KL206902 KL206932 KL206979 KL206995 KL206996 KL207002 KFV73492.1 KFV73612.1 KFV74069.1 KFV77013.1 KFV78397.1 KFV80821.1 KFV81678.1 KFV81842.1 KFV82263.1 KFV82322.1 KFV82483.1 KFV83356.1 KFV85496.1 KFV86827.1 KFV87138.1 KFV87825.1 KFV88414.1 KFV88462.1 KFV88705.1 KL206410 KFV82084.1 KL206549 KFV83237.1 KL205926 KFV77067.1 KFV73601.1 KL206733 KFV85403.1 KL205665 KFV72673.1 KL205888 KFV76262.1 KL206562 KFV83655.1 KFV81119.1 KL206071 KFV78608.1 KFV83965.1 QRBI01000220 RMB92612.1 KL206129 KFV79085.1 KL206190 KFV79766.1 QRBI01000131 RMC03475.1 MRZV01001153 PIK40187.1 KL206600 KFV84076.1 KL207003 KFV88757.1 KL206694 KFV84978.1 KL206630 KFV84296.1 KL206188 KFV79735.1 QRBI01000111 RMC10945.1 KL206273 KFV80807.1 QRBI01000093 RMC21953.1 QRBI01000126 RMC04361.1 KL206086 KFV78804.1 KFV88760.1 QRBI01000106 RMC12940.1 KL205829 KFV75232.1 KL205810 KFV74798.1 KL206513 KFV82948.1 KL206811 KFV86001.1 KL206812 KFV86049.1 KFV81420.1 KL205877 KFV75915.1 RMC21288.1 KL206301 KFV80966.1 KFV76460.1 KL206514 KFV82949.1 KL205740 KFV73754.1 KL206036 KFV78283.1 KL217824 KFO99526.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR013083 Znf_RING/FYVE/PHD
IPR036987 SRA-YDG_sf
IPR018957 Znf_C3HC4_RING-type
IPR015947 PUA-like_sf
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR003105 SRA_YDG
IPR031986 GD_N
IPR008936 Rho_GTPase_activation_prot
IPR036865 CRAL-TRIO_dom_sf
IPR039360 Ras_GTPase
IPR001936 RasGAP_dom
IPR011993 PH-like_dom_sf
IPR023152 RasGAP_CS
IPR001251 CRAL-TRIO_dom
IPR029479 Nitroreductase
IPR000415 Nitroreductase-like
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR013083 Znf_RING/FYVE/PHD
IPR036987 SRA-YDG_sf
IPR018957 Znf_C3HC4_RING-type
IPR015947 PUA-like_sf
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR003105 SRA_YDG
IPR031986 GD_N
IPR008936 Rho_GTPase_activation_prot
IPR036865 CRAL-TRIO_dom_sf
IPR039360 Ras_GTPase
IPR001936 RasGAP_dom
IPR011993 PH-like_dom_sf
IPR023152 RasGAP_CS
IPR001251 CRAL-TRIO_dom
IPR029479 Nitroreductase
IPR000415 Nitroreductase-like
SUPFAM
CDD
ProteinModelPortal
Q5KTM5
Q6UV17
A0A2H1WAR9
A0A2H1VFZ3
B9WPT5
T2M6M7
+ More
A0A2H1V2F4 J9GT67 A0A2H1WCG9 A0A3D5S1K1 A0A2H1VUV7 A0A0P4VXU0 A0A2H1WY65 T2MGM5 A0A1Y1S3P2 A0A2H1W7F5 A0A1E1XGQ7 A0A093I968 A0A093H5S5 A0A093JFI6 A0A093HMV3 A0A093GU34 A0A093IBP9 A0A093JL05 A0A093H7K6 A0A093HHB4 A0A093HVG8 A0A093JHV8 A0A093HF89 A0A093HFB7 A0A093HCT5 A0A093HJX0 A0A093HT80 A0A093HYW2 A0A093HBM3 A0A093JC36 A0A093HPP7 A0A093HNU6 A0A093HPY1 A0A093HPA2 A0A093I7Q5 A0A093GVA5 A0A093HMV9 A0A093J7J9 A0A093HGI5 A0A093JU31 A0A093HW56 A0A093HDI4 A0A093GW31 A0A093HRW3 A0A093GQT0 A0A093HB81 A0A093HVT9 A0A093JR35 A0A093H7L9 A0A093HWQ4 A0A3M0IVW1 A0A093HHZ8 A0A093HDI8 A0A3M0JRY7 A0A2G8JWN9 A0A093HQY8 A0A093IC93 A0A093HQN0 A0A093HRM3 A0A093HJW5 A0A3M0KD23 A0A093HP87 A0A3M0L9G9 A0A3M0JVZ5 A0A093JJ50 A0A093IA31 A0A3M0KPI8 A0A093H881 A0A093GWX3 A0A093HJY5 A0A093HWI0 A0A093K6G3 A0A093HPR5 A0A093H950 A0A3M0L7E2 A0A093HGX5 A0A093HAN0 A0A093JWX0 A0A093J469 A0A093H6R6 A0A091HUB5
A0A2H1V2F4 J9GT67 A0A2H1WCG9 A0A3D5S1K1 A0A2H1VUV7 A0A0P4VXU0 A0A2H1WY65 T2MGM5 A0A1Y1S3P2 A0A2H1W7F5 A0A1E1XGQ7 A0A093I968 A0A093H5S5 A0A093JFI6 A0A093HMV3 A0A093GU34 A0A093IBP9 A0A093JL05 A0A093H7K6 A0A093HHB4 A0A093HVG8 A0A093JHV8 A0A093HF89 A0A093HFB7 A0A093HCT5 A0A093HJX0 A0A093HT80 A0A093HYW2 A0A093HBM3 A0A093JC36 A0A093HPP7 A0A093HNU6 A0A093HPY1 A0A093HPA2 A0A093I7Q5 A0A093GVA5 A0A093HMV9 A0A093J7J9 A0A093HGI5 A0A093JU31 A0A093HW56 A0A093HDI4 A0A093GW31 A0A093HRW3 A0A093GQT0 A0A093HB81 A0A093HVT9 A0A093JR35 A0A093H7L9 A0A093HWQ4 A0A3M0IVW1 A0A093HHZ8 A0A093HDI8 A0A3M0JRY7 A0A2G8JWN9 A0A093HQY8 A0A093IC93 A0A093HQN0 A0A093HRM3 A0A093HJW5 A0A3M0KD23 A0A093HP87 A0A3M0L9G9 A0A3M0JVZ5 A0A093JJ50 A0A093IA31 A0A3M0KPI8 A0A093H881 A0A093GWX3 A0A093HJY5 A0A093HWI0 A0A093K6G3 A0A093HPR5 A0A093H950 A0A3M0L7E2 A0A093HGX5 A0A093HAN0 A0A093JWX0 A0A093J469 A0A093H6R6 A0A091HUB5
Ontologies
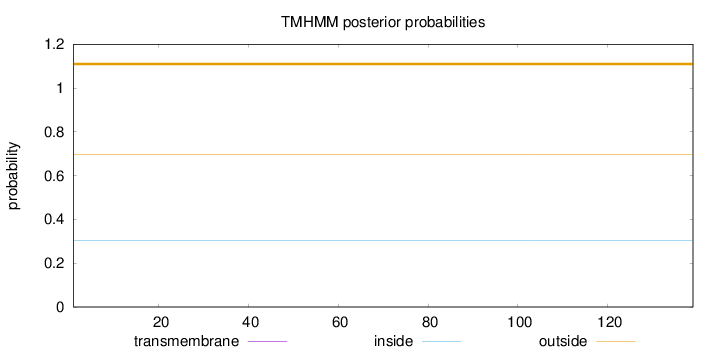
Topology
Subcellular location
Nucleus
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000690000000000001
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.30489
outside
1 - 139
Population Genetic Test Statistics
Pi
31.558901
Theta
28.821086
Tajima's D
0.573091
CLR
1.045134
CSRT
0.549772511374431
Interpretation
Uncertain
